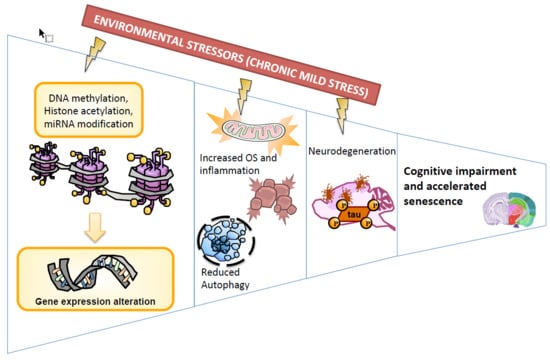
Chronic Mild Stress Modified Epigenetic Mechanisms Leading to Accelerated Senescence and Impaired Cognitive Performance in Mice
Abstract
:1. Introduction
2. Results
2.1. Epigenetic Modulation Triggers Compacted Chromatin after Chronic Mild Stress
2.2. Chronic Mild Stress Attenuates Antioxidant Defence Mechanisms
2.3. Inflammatory Activation is Induced after Application of Chronic Mild Stress
2.4. Chronic Mild Stress Modulates APP Processing as Well as Alters Microtubule Stabilization Through Hyperphosphorylation of Tau Protein
2.5. Chronic Mild Stress Promotes Autophagy Activation
2.6. Anxiety-Like Behaviour, Memory Decline and Poorer Cognitive Abilities After Chronic Mild Stress
3. Discussion
4. Materials and Methods
4.1. Animals and Chronic Mild Stress Procedure
4.2. Glucose Tolerance Test
4.3. Behavioural and Cognitive Test
4.3.1. Open Field Test (OFT)
4.3.2. Novel Object Recognition Test (NORT)
4.3.3. Morris Water Maze (MWM)
4.4. Immunodetection Experiments
4.4.1. Brain Processing
4.4.2. Western Blotting
4.5. RNA Extraction and Gene Expression Determination by q-PCR
4.6. Global DNA Methylation and Hydroxymethylation Determination
4.7. miRNA Expression Array and Validation by Single Real-Time PCR
4.8. Oxidative Stress Determination
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cristòfol, R.; Porquet, D.; Corpas, R.; Coto-Montes, A.; Serret, J.; Camins, A.; Pallàs, M.; Sanfeliu, C. Neurons from senescence-accelerated SAMP8 mice are protected against frailty by the Sirtuin 1 promoting agents melatonin and resveratrol. J. Pineal Res. 2012, 52, 271–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harman, M.F.; Martín, M.G. Epigenetic mechanisms related to cognitive decline during aging. J. Neurosci. Res. 2019, 98, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, R.; Bras, J. The age factor in Alzheimer’s disease. Genome Med. 2015, 7, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griñan-Ferré, C.; Puigoriol-Illamola, D.; Palomera-Ávalos, V.; Pérez-Cáceres, D.; Companys-Alemany, J.; Camins, A.; Ortuño-Sahagún, D.; Rodrigo, M.T.; Pallàs, M. Environmental Enrichment Modified Epigenetic Mechanisms in SAMP8 Mouse Hippocampus by Reducing Oxidative Stress and Inflammaging and Achieving Neuroprotection. Front. Aging Neurosci. 2016, 8, 26071. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Kennedy, P.; Nestler, E. Epigenetics of the Depressed Brain: Role of Histone Acetylation and Methylation. Neuropsychopharmacol 2013, 38, 124–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turecki, G.; Meaney, M.J. Effects of the Social Environment and Stress on Glucocorticoid Receptor Gene Methylation: A Systematic Review. Biol. Psychiatry 2016, 79, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Sandi, C. Stress and Cognition. WIREs Cogn. Sci. 2013, 4, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.E.; Ko, S.Y.; Jo, S.; Jo, H.-R.; Han, J.; Kim, Y.-S.; Son, H. Downregulation of SIRT2 by Chronic Stress Reduces Expression of Synaptic Plasticity-related Genes through the Upregulation of Ehmt2. Exp. Neurobiol. 2019, 28, 537–546. [Google Scholar] [CrossRef]
- Borcel, E.; Pérez-Álvarez, L.; Herrero, A.I.; Brionne, T.; Varea, E.; Berezin, V.; Bock, E.; Sandi, C.; Venero, C. Chronic stress in adulthood followed by intermittent stress impairs spatial memory and the survival of newborn hippocampal cells in aging animals: Prevention by FGL, a peptide mimetic of neural cell adhesion molecule. Behav. Pharmacol. 2008, 19, 41–49. [Google Scholar] [CrossRef]
- Rahman, M.M.; Callaghan, C.K.; Kerskens, C.M.; Chattarji, S.; O’Mara, S.M. Early hippocampal volume loss as a marker of eventual memory deficits caused by repeated stress. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Jett, J.D.; Bulin, S.E.; Hatherall, L.C.; McCartney, C.M.; Morilak, D.A. Deficits in cognitive flexibility induced by chronic unpredictable stress are associated with impaired glutamate neurotransmission in the rat medial prefrontal cortex. Neuroscience 2017, 346, 284–297. [Google Scholar] [CrossRef] [Green Version]
- Justice, N.J. The relationship between stress and Alzheimer’s disease. Neurobiol. Stress 2018, 8, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Bisht, K.; Sharma, K.; Tremblay, M. Chronic stress as a risk factor for Alzheimer’s disease: Roles of microglia-mediated synaptic remodeling, inflammation, and oxidative stress. Neurobiol. Stress 2018, 9, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Griñan-Ferré, C.; Corpas, R.; Puigoriol-Illamola, D.; Palomera-Avalos, V.; Sanfeliu, C.; Pallàs, M. Understanding Epigenetics in the Neurodegeneration of Alzheimer’s Disease: SAMP8 Mouse Model. J. Alzheimers Dis. 2018, 62, 943–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, H.Y.; Kim, D.H.; Lee, E.K.; Chung, K.W.; Chung, S.; Lee, B.; Seo, A.Y.; Chung, J.H.; Jung, Y.S.; Im, E.; et al. Redefining Chronic Inflammation in Aging and Age-Related Diseases: Proposal of the Senoinflammation Concept. Aging Dis. 2019, 10, 367–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, K.S.; Smith, C. Ageing-Associated Oxidative Stress and Inflammation Are Alleviated by Products from Grapes. Oxidative Med. Cell. Longev. 2016, 2016, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Escobar, K.A.; Cole, N.H.; Mermier, C.M.; VanDusseldorp, T.A. Autophagy and aging: Maintaining the proteome through exercise and caloric restriction. Aging Cell 2019, 18. [Google Scholar] [CrossRef] [Green Version]
- Gouras, G.K. Aging, Metabolism, Synaptic Activity, and Aβ in Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 185. [Google Scholar] [CrossRef] [Green Version]
- Puigoriol-Illamola, D.; Griñan-Ferré, C.; Vasilopoulou, F.; Leiva, R.; Vázquez, S.; Pallàs, M. 11β-HSD1 Inhibition by RL-118 Promotes Autophagy and Correlates with Reduced Oxidative Stress and Inflammation, Enhancing Cognitive Performance in SAMP8 Mouse Model. Mol. Neurobiol. 2018, 55, 8904–8915. [Google Scholar] [CrossRef]
- Martinez-Lopez, N.; Athonvarangkul, D.; Singh, R. Autophagy and aging. Single Mol. Single Cell Seq. 2015, 847, 73–87. [Google Scholar]
- Takeda, T.; Hosokawa, M.; Higuchi, K. Senescence-Accelerated Mouse (SAM): A Novel Murine Model of Accelerated Senescence. J. Am. Geriatr. Soc. 1991, 39, 911–919. [Google Scholar] [CrossRef]
- Wang, J.; Yuan, J.; Pang, J.; Ma, J.; Han, B.; Geng, Y.; Shen, L.; Wang, H.; Ma, Q.; Wang, Y.; et al. Effects of Chronic Stress on Cognition in Male SAMP8 Mice. Cell. Physiol. Biochem. 2016, 39, 1078–1086. [Google Scholar] [CrossRef]
- Foster, J.A.; Rinaman, L.; Cryan, J.F. Stres & the gut-brain axis: Regulation by the microbiome. Neuribiol. Stress 2017, 7, 124–136. [Google Scholar]
- Watson, J.D.; Baker, T.A.; Gann, A.; Levine, M.; Losik, R. Molecular Biology of the Gene, 7th ed.; Pearson/CSH Press: Boston, MA, USA, 2014. [Google Scholar]
- Turner, B.M. Histone acetylation and an epigenetic code. BioEssays 2000, 22, 836–845. [Google Scholar] [CrossRef]
- Peleg, S.; Sananbenesi, F.; Zovoilis, A.; Burkhardt, S.; Bahari-Javan, S.; Agis-Balboa, R.C.; Cota, P.; Wittnam, J.L.; Gogol-Doering, A.; Opitz, L.; et al. Altered Histone Acetylation Is Associated with Age-Dependent Memory Impairment in Mice. Science 2010, 328, 753–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuadrado-Tejedor, M.; Pérez-González, M.; García-Muñoz, C.; Muruzabal, D.; García-Barroso, C.; Rabal, O.; Segura, V.; Sánchez-Arias, J.A.; Oyarzabal, J.O.; Garcia-Osta, A. Taking Advantage of the Selectivity of Histone Deacetylases and Phosphodiesterase Inhibitors to Design Better Therapeutic Strategies to Treat Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 149. [Google Scholar] [CrossRef] [Green Version]
- Cosãn-Tomã¡s, M.; Alvarez-Lã³Pez, M.J.; Sanchez-Roige, S.; Lalanza, J.F.; Bayod, S.; Sanfeliu, C.; Pallas, M.; Escorihuela, R.M.; Kaliman, P. Epigenetic alterations in hippocampus of SAMP8 senescent mice and modulation by voluntary physical exercise. Front. Aging Neurosci. 2014, 6, 51. [Google Scholar] [CrossRef]
- Herskovits, A.Z.; Guarente, L. SIRT1 in neurodevelopment and brain senescence. Neuron 2014, 81, 471–483. [Google Scholar] [CrossRef] [Green Version]
- Pallàs, M. Senescence-Accelerated Mice P8: A Tool to Study Brain Aging and Alzheimer’s Disease in a Mouse Model. Cell Biol. 2012, 12, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Fourcade, S.; Outeiro, T.F.; Pujol, A. SIRT2 in age-related neurodegenerative disorders. Aging 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.I.; Nirzhor, S.S.R.; Akter, R. A Review of the Recent Advances Made with SIRT6 and its Implications on Aging Related Processes, Major Human Diseases, and Possible Therapeutic Targets. Biomolecules 2018, 8, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jesko, H.; Wencel, P.; Strosznajder, R.P.; Strosznajder, J.B. Sirtuins and Their Roles in Brain Aging and Neurodegenerative Disorders. Neurochem. Res. 2017, 42, 876–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Zhang, J.; Chen, R.; Wang, L.; Li, B.; Cheng, H.; Duan, X.; Zhu, H.; Wei, W.; Li, J.; et al. Dissecting the precise role of H3K9 methylation in crosstalk with DNA maintenance methylation in mammals. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, J.M.; Perrin, R.M.; Dagenais, T.R.T.; Keller, N.P. H3K9 Methylation Regulates Growth and Development in Aspergillus fumigatus. Eukaryot. Cell 2008, 7, 2052–2060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossetto, D.; Avvakumov, N.; Côte, J. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef] [Green Version]
- Fetahu, I.S.; Ma, D.; Rabidou, K.; Argueta, C.; Smith, M.; Liu, H.; Wu, F.; Shi, Y.G. Epigenetic signatures of methylated DNA cytosine in Alzheimer’s disease. Sci. Adv. 2019, 5. [Google Scholar] [CrossRef] [Green Version]
- Sherwani, S.I.; Khan, H.A. Role of 5-hydroxymethylcytosine in neurodegeneration. Gene 2015, 570, 17–24. [Google Scholar] [CrossRef]
- Yokoyama, A.S.; Rutledge, J.C.; Medici, V. DNA methylation alterations in Alzheimer’s disease. Environ. Epigenet. 2017, 3. [Google Scholar] [CrossRef]
- Chestnut, B.A.; Chang, Q.; Price, A.; Lesuisse, C.; Wong, M.; Martin, L.J. Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 2011, 31, 16619–16636. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, D.G.; Nalls, M.A.; Gibbs, J.R.; Arepalli, S.; Van Der Brug, M.; Chong, S.; Moore, M.; Longo, D.L.; Cookson, M.R.; Traynor, B.J.; et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 2011, 20, 1164–1172. [Google Scholar] [CrossRef] [Green Version]
- Llorens-Martín, M.; Jurado, J.; Hernández, F.; Avila, J. GSK-3β, a pivotal kinase in Alzheimer disease. Front. Mol. Neurosci. 2014, 7. [Google Scholar] [CrossRef]
- Ly, P.T.T.; Wu, Y.; Zou, H.; Wang, R.; Zhou, W.; Kinoshita, A.; Zhang, M.; Yang, Y.; Cai, F.; Woodgett, J.; et al. Inhibition of GSK-3β-mediated BACE1 expression reduces Alzheimer-associated phenotypes. J. Clin. Investig. 2013, 123, 224–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reichenbach, N.; Delekate, A.; Plescher, M.; Schmitt, F.; Krauss, S.; Blank, N.; Halle, A.; Petzold, G.C. Inhibition of Stat3-mediated astrogliosis ameliorates pathology in an Alzheimer’s disease model. EMBO Mol. Med. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.; McCarthy, V.L. Stress and anxiety among nursing students: A review of intervention strategies in literature between 2009 and 2015. Nurse Educ. Pract. 2017, 22, 21–29. [Google Scholar] [CrossRef]
- Stilling, R.M.; Fischer, A. The role of histone acetylation in age-associated memory impairment and Alzheimer’s disease. Neurobiol. Learn. Mem. 2011, 96, 19–26. [Google Scholar] [CrossRef]
- Currais, A.; Prior, M.; Lo, D.; Jolivalt, C.; Schubert, D.; Maher, P. Diabetes exacerbates amyloid and neurovascular pathology in aging-accelerated mice. Aging Cell 2012, 11, 1017–1026. [Google Scholar] [CrossRef] [Green Version]
- Griñán-Ferré, C.; Pérez-Cáceres, D.; Gutiérrez-Zetina, S.M.; Camins, A.; Palomera-Ávalos, V.; Ortuño-Sahagun, D.; Rodrigo, M.T.; Pallàs, M. Environmental Enrichment Improves Behavior, Cognition, and Brain Functional Markers in Young Senescence-Accelerated Prone Mice (SAMP8). Mol. Neurobiol. 2015, 53, 2435–2450. [Google Scholar] [CrossRef]
- Puigoriol-Illamola, D.; Leiva, R.; Vázquez-Carrera, M.; Vázquez, S.; Griñán-Ferré, C.; Pallàs, M. 11β-HSD1 Inhibition Rescues SAMP8 Cognitive Impairment Induced by Metabolic Stress. Mol. Neurobiol. 2019, 57, 551–565. [Google Scholar] [CrossRef]
- Vorhees, C.V.; Williams, M.T. Morris water maze: Procedures for assessing spatial and related forms of learning and memory. Nat. Protoc. 2006, 1, 848–858. [Google Scholar] [CrossRef] [Green Version]











© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puigoriol-Illamola, D.; Martínez-Damas, M.; Griñán-Ferré, C.; Pallàs, M. Chronic Mild Stress Modified Epigenetic Mechanisms Leading to Accelerated Senescence and Impaired Cognitive Performance in Mice. Int. J. Mol. Sci. 2020, 21, 1154. https://doi.org/10.3390/ijms21031154
Puigoriol-Illamola D, Martínez-Damas M, Griñán-Ferré C, Pallàs M. Chronic Mild Stress Modified Epigenetic Mechanisms Leading to Accelerated Senescence and Impaired Cognitive Performance in Mice. International Journal of Molecular Sciences. 2020; 21(3):1154. https://doi.org/10.3390/ijms21031154
Chicago/Turabian StylePuigoriol-Illamola, Dolors, Mirna Martínez-Damas, Christian Griñán-Ferré, and Mercè Pallàs. 2020. "Chronic Mild Stress Modified Epigenetic Mechanisms Leading to Accelerated Senescence and Impaired Cognitive Performance in Mice" International Journal of Molecular Sciences 21, no. 3: 1154. https://doi.org/10.3390/ijms21031154
APA StylePuigoriol-Illamola, D., Martínez-Damas, M., Griñán-Ferré, C., & Pallàs, M. (2020). Chronic Mild Stress Modified Epigenetic Mechanisms Leading to Accelerated Senescence and Impaired Cognitive Performance in Mice. International Journal of Molecular Sciences, 21(3), 1154. https://doi.org/10.3390/ijms21031154