Interactions of Whey Proteins with Metal Ions
Abstract
:1. Introduction
1.1. β-Lactoglobulin
1.2. α-Lactalbumin
1.3. Lactoferrin
2. Interaction of Whey Proteins with Metals
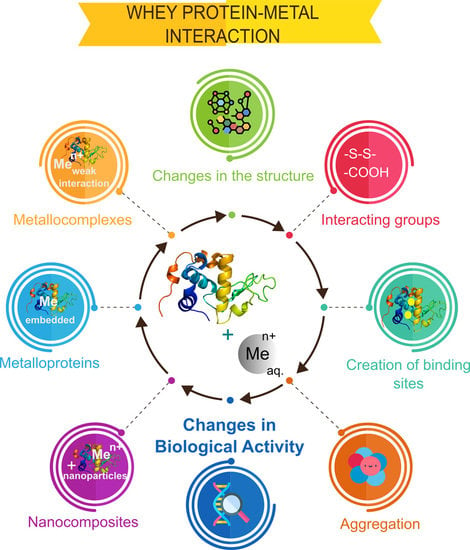
2.1. Nature of the Metal–Protein Interaction
2.2. Analytical techniques for separation and analysis of whey proteins
2.3. Analytical Techniques for Studies of Interactions of Whey Proteins with Metal Ions
2.3.1. Mass Spectrometry
2.3.2. Spectroscopic Techniques
2.3.3. Microscopic Techniques
2.3.4. Complementarity of MALDI- and NALDI-TOF-MS for Metal–Protein Interactions Studies
3. Implications of Whey Protein–Metal Interactions in Food and Nutraceuticals
3.1. Changes in Bioactivity after Metal Interactions
3.2. Consequences of Changes in Biological Activity in Food and Nutraceuticals
4. Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-FU | 5-Fluorouracil |
| α-LA | α-lactalbumin |
| β-LG | β-lactoglobulin |
| AFM | Atomic Force Microscopy |
| BSA | Bovine Serum Albumin |
| CE | Capillary Electrophoresis |
| CD | Circular Dichroism |
| DAD | Diode Array Detector |
| DFT | Density Functional Theory |
| DLS | Dynamic Light Scattering |
| DLVO | Derjagin–Landau–Verwey–Overbeek theory of intraparticle interactions |
| DSC | Differential Scanning Calorimetry |
| DTNB | Dithio(bis)-p-nitrobenzoate |
| EDX | Energy Dispersive X-ray Spectroscopy |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| ESI-MS/MS | Electrospray Ionization Tandem Mass Spectrometry |
| FAAS | Flame Atomic Absorption Spectrometry |
| FE-SEM | Field Emission Scanning Electron Microscopes |
| FRET | Förster Resonance Energy Transfer |
| FT-ICR | Fourier transform ion cyclotron resonance |
| FPLC | Fast Protein Liquid Chromatography |
| HBM | Human Breast Milk |
| HPCEC | High Performance Cation Exchange Chromatography |
| HPIMAC | High Performance Immobilized Metal Ion Affinity Chromatography |
| HPLC | High Performance Liquid Chromatography |
| HPSEC | High Performance Size Exclusion Chromatography |
| HSA | Human Serum Albumin |
| IG | Immunoglobulin |
| ICP-MS | Inductively Coupled Plasma Mass Spectrometry |
| IMAC | Immobilized Metal Affinity Chromatography |
| ITC | Isothermal Titration Calorimetry |
| iTRAQ | Isobaric Tags for Relative and Absolute Quantization |
| LC-ESI-MS | Liquid Chromatography Electrospray Ionization-Mass Spectrometry |
| LTF | Lactoferrin |
| LP | Lactoperoxidase |
| MALDI-TOF MS | Matrix-Assisted Laser Desorption Ionization technique coupled to Time-of-Flight Mass Spectrometry |
| MCE | Microchip Electrophoresis |
| MD | Molecular Dynamics |
| NALDI | Nano-Assisted Laser Desorption Ionization |
| NEM | N-ethylmaleimide |
| NMR | Nuclear Magnetic Resonance |
| PDA | Photodiode Array Detector |
| PTMs | Posttranslational Modifications |
| ROS | Reactive Oxygen Species |
| RS | Raman Spectroscopy |
| SDS-PAGE | Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis |
| SEC | Size Exclusion Chromatography |
| SEC-ICP-MS | Size Exclusion Chromatography-Inductively Coupled Plasma-Mass Spectrometry |
| SERS | Surface Enhanced Raman Spectroscopy |
| SEM | Scanning Electron Microscope |
| SEP buffer | Separating Milk Protein Buffer |
| TEM | Transmission Electron Microscope |
| TPS Buffer | Total Protein Solubilization Buffer |
| TG-DTA | Thermogravimetry/Differential Thermal Analysis |
| UV-Vis | Ultraviolet/ Visible Spectroscopy |
| WPC | Whey Protein Concentrate |
| WPH | Whey Protein Hydrolysate |
| WPI | Whey Protein Isolate |
| XPS | X-ray Photoelectron Spectroscopy |
References
- Madureira, A.R.; Pereira, C.I.; Gomes, A.M.P.; Pintado, M.E.; Xavier Malcata, F. Bovine whey proteins—Overview on their main biological properties. Food Res. Int. 2007, 40, 1197–1211. [Google Scholar] [CrossRef]
- Chatterton, D.E.W.; Smithers, G.; Roupas, P.; Brodkorb, A. Bioactivity of b -lactoglobulin and a -lactalbumin — Technological implications for processing. Int. Dairy J. 2006, 16, 1229–1240. [Google Scholar] [CrossRef]
- Corrochano, A.R.; Buckin, V.; Kelly, P.M.; Giblin, L. Invited review: Whey proteins as antioxidants and promoters of cellular antioxidant pathways. J. Dairy Sci. 2018, 101, 4747–4761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godovac-Zimmermann, J.; Krause, I.; Baranyi, M.; Fischer-Frühholz, S.; Juszczak, J.; Erhardt, G.; Buchberger, J.; Klostertneyer, H. Isolation and Rapid Sequence Characterization of Two Novel Bovine 13-Lactoglobulins I and J. J. Protein Chem. 1996, 15, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lu, Y.; Xu, H.; Lin, D.; He, Z.; Wu, H.; Liu, L.; Wang, Z. Reducing the allergenic capacity of β -lactoglobulin by covalent conjugation with dietary polyphenols. Food Chem. 2018, 256, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Mensi, A.; Choiset, Y.; Rabesona, H.; Haertlé, T.; Borel, P.; Chobert, J.-M. Interactions of β-Lactoglobulin Variants A and B with Vitamin A. Competitive Binding of Retinoids and Carotenoids. J. Agric. Food Chem. 2013, 61, 4114–4119. [Google Scholar] [CrossRef]
- Qin, B.Y.; Bewley, M.C.; Creamer, L.K.; Baker, H.M.; Baker, E.N.; Jameson, G.B. Structural Basis of the Tanford transition of Bovine beta-Lactoglobulin. Biochemistry 1998, 37, 14014–14023. [Google Scholar] [CrossRef]
- Ma, S.; Wang, C.; Guo, M. Changes in structure and antioxidant activity of β-lactoglobulin by ultrasound and enzymatic treatment. Ultrason. Sonochem. 2018, 43, 227–236. [Google Scholar] [CrossRef]
- Sakai, K.; Sakurai, K.; Sakai, M.; Hoshino, M.; Goto, Y. Conformation and stability of thiol-modified bovine beta-lactoglobulin. Protein Sci. 2000, 9, 1719–1729. [Google Scholar]
- Enomoto, H.; Li, C.P.; Morizane, K.; Ibrahim, H.R.; Sugimoto, Y.; Ohki, S.; Ohtomo, H.; Aoki, T. Glycation and Phosphorylation of β-Lactoglobulin by Dry-Heating: Effect on Protein Structure and Some Properties. J. Agric. Food Chem. 2007, 55, 2392–2398. [Google Scholar] [CrossRef]
- Pinto, M.S.; Léonil, J.; Henry, G.; Cauty, C.; Carvalho, A.F.; Bouhallab, S. Heating and glycation of β -lactoglobulin and β -casein: Aggregation and in vitro digestion. Food Res. Int. 2014, 55, 70–76. [Google Scholar] [CrossRef]
- Mann, M.; Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003, 21, 255–261. [Google Scholar] [CrossRef] [PubMed]
- de Wit, J.N. Thermal behaviour of bovine β-lactoglobulin at temperatures up to 150 °C. A review. Trends Food Sci. Technol. 2009, 20, 27–34. [Google Scholar] [CrossRef]
- Liu, H.C.; Chen, W.L.; Mao, S.J.T. Antioxidant Nature of Bovine Milk β -Lactoglobulin. J. Dairy Sci. 2007, 90, 547–555. [Google Scholar] [CrossRef]
- Wijayanti, H.B.; Oh, H.E.; Sharma, R.; Deeth, H.C. Reduction of aggregation of β-lactoglobulin during heating by dihydrolipoic acid. J. Dairy Res. 2013, 80, 383–389. [Google Scholar] [CrossRef]
- Medrano, A.; Abirached, C.; Panizzolo, L.; Moyna, P.; Añón, M.C. The effect of glycation on foam and structural properties of β-lactoglobulin. Food Chem. 2009, 113, 127–133. [Google Scholar] [CrossRef]
- Aich, R.; Batabyal, S.; Joardar, S.N. Isolation and purification of beta-lactoglobulin from cow milk. Vet. World 2015, 8, 621–624. [Google Scholar] [CrossRef] [Green Version]
- Hogarth, C.J.; Fitzpatrick, J.L.; Nolan, A.M.; Young, F.J.; Pitt, A.; Eckersall, P.D. Differential protein composition of bovine whey: A comparison of whey from healthy animals and from those with clinical mastitis. Proteomics 2004, 4, 2094–2100. [Google Scholar] [CrossRef]
- Svensson, M.; Sabharwal, H.; Håkansson, A.; Mossberg, A.K.; Lipniunas, P.; Leffler, H.; Svanborg, C.; Linse, S. Molecular characterization of alpha-lactalbumin folding variants that induce apoptosis in tumor cells. J. Biol. Chem. 1999, 274, 6388–6396. [Google Scholar] [CrossRef] [Green Version]
- Froehlich, J.W.; Dodds, E.D.; Barboza, M.; Mcjimpsey, E.L.; Richard, R.; Francis, J.; An, H.J.; Freeman, S.; German, J.B.; Lebrilla, C.B. Glycoprotein Expression in Human Milk During Lactation. J. Agric. Food Chem. 2010, 58, 6440–6448. [Google Scholar] [CrossRef] [Green Version]
- Fong, B.Y.; Norris, C.S.; Palmano, K.P. Fractionation of bovine whey proteins and characterisation by proteomic techniques. Int. Dairy J. 2008, 18, 23–46. [Google Scholar] [CrossRef]
- Eigel, W.N.; Butler, J.E.; Ernstrom, C.A.; Farrell, H.M.; Harwalkar, V.R.; Jenness, R.; Whitney, R.M.L. Nomenclature of Proteins of Cow’s Milk: Fifth Revision. J. Dairy Sci. 1984, 67, 1599–1631. [Google Scholar] [CrossRef]
- Mercadante, D.; Melton, L.D.; Norris, G.E.; Loo, T.S.; Williams, M.A.K.; Dobson, R.C.J.; Jameson, G.B. Bovine β-Lactoglobulin is dimeric under imitative physiological conditions: Dissociation equilibrium and rate constants over the pH range of 2.5-7.5. Biophys. J. 2012, 103, 303–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, R.V.; Butkowski, R.J.; Ebner, K.E.; Hamilton, J.W. Amino Acid Sequence of Rat α-Lactalbumin: A Unique α-Lactalbumin. Biochemistry 1982, 21, 1479–1482. [Google Scholar] [CrossRef]
- Layman, D.K.; Lönnerdal, B.; Fernstrom, J.D. Applications for a-lactalbumin in human nutrition. Nutr. Rev. 2018, 76, 444–460. [Google Scholar] [CrossRef]
- Noyelle, K.; Van Dael, H. Kinetics of conformational changes induced by the binding of various metal ions to bovine α-lactalbumin. J. Inorg. Biochem. 2002, 88, 69–76. [Google Scholar] [CrossRef]
- Chandra, N.; Brew, K.; Ravi, A.K. Structural Evidence for the Presence of a Secondary Calcium Binding Site in Human R-Lactalbumin. Biochemistry 1998, 37, 4767–4772. [Google Scholar] [CrossRef]
- Kim, S.; Baum, J. Electrostatic interactions in the acid denaturation of α-lactalbumin determined by NMR. Protein Sci. 1998, 7, 1930–1938. [Google Scholar] [CrossRef]
- Veprintsev, D.B.; Permyakov, S.E.; Permyakov, E.A.; Rogov, V.V.; Cawthern, K.M.; Berliner, L.J. Cooperative thermal transitions of bovine and human apo-α-lactalbumins: Evidence for a new intermediate state. FEBS Lett. 1997, 412, 625–628. [Google Scholar] [CrossRef] [Green Version]
- Shinozaki, R.; Iwaoka, M. Effects of metal ions, temperature, and a denaturant on the oxidative folding pathways of bovine α-lactalbumin. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [Green Version]
- Kronman, M.J.; Sinha, S.K.; Brew, K. Characteristics of the binding of Ca2+ and other divalent metal ions to bovine alpha-lactalbumin. J. Biol. Chem. 1981, 256, 8582–8587. [Google Scholar] [PubMed]
- Wehbi, Z.; Pérez, M.D.; Sánchez, L.; Pocoví, C.; Barbana, C.; Calvo, M. Effect of heat treatment on denaturation of bovine α-lactalbumin: Determination of kinetic and thermodynamic parameters. J. Agric. Food Chem. 2005, 53, 9730–9736. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, E.A.; Berliner, L.J. α-Lactalbumin: structure and function. FEBS Lett. 2000, 473, 269–274. [Google Scholar] [CrossRef] [Green Version]
- Fujita-Yamaguchi, Y. Affinity chromatography of native and recombinant proteins from receptors for insulin and IGF-I to recombinant single chain antibodies. Front. Endocrinol. (Lausanne). 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Bushmarina, N.A.; Blanchet, C.E.; Vernier, G.; Forge, V. Cofactor effects on the protein folding reaction: Acceleration of α-lactalbumin refolding by metal ions. Protein Sci. 2006, 15, 659–671. [Google Scholar] [CrossRef] [Green Version]
- Atri, M.S.; Saboury, A.A.; Moosavi-Movahedi, A.A.; Kavousi, K.; Ariaeenejad, S. Effects of zinc binding on the structure and thermal stability of camel alpha-lactalbumin. J. Therm. Anal. Calorim. 2015, 120, 481–488. [Google Scholar] [CrossRef]
- García-Montoya, I.A.; Cendón, T.S.; Arévalo-Gallegos, S.; Rascón-Cruz, Q. Lactoferrin a multiple bioactive protein: An overview. Biochim. Biophys. Acta 2012, 1820, 226–236. [Google Scholar] [CrossRef]
- Wei, Z.; Nishimura, T.; Yoshida, S. Presence of a Glycan at a Potential N-Glycosylation Site, Asn-281, of Bovine Lactoferrin. J. Dairy Sci. 2000, 83, 683–689. [Google Scholar] [CrossRef]
- Redwan, E.M.; Uversky, V.N.; El-Fakharany, E.M.; Al-Mehdar, H. Potential lactoferrin activity against pathogenic viruses. Comptes Rendus Biol. 2014, 337, 581–595. [Google Scholar] [CrossRef]
- Atkins, P.W.; Overton, T.L.; Rourke, J.P.; Weller, M.T.; Armstrong, F. Inorganic Chemistry, Fifth Edition; W. H. Freeman and Company: New York, NY, USA, 2010; ISBN 978-1-42-921820-7. [Google Scholar]
- Housecroft, C.E.; Sharpe, A.G. Inorganic Chemistry Third Edition; Pearson Education: Edinburgh Gate, UK, 2008; ISBN 978-0-13-175553-6. [Google Scholar]
- Baker, E.N. Structure and reactivity of transferrins. Adv. Inorg. Chem. 1994, 41, 389–463. [Google Scholar]
- Mesonjesi, I. Are extrinsic black stains of teeth iron-saturated bovine lactoferrin and a sign of iron deficient anemia or iron overload? Med. Hypotheses 2012, 79, 219–221. [Google Scholar] [CrossRef]
- Lestas, A.N. The Effect of pH upon Human Transferrin: Selective Labelling of the Two Iron-binding Sites. Br. J. Haematol. 1976, 32, 341–350. [Google Scholar] [CrossRef]
- Valenti, P.; Antonini, G. Lactoferrin: An important host defence against microbial and viral attack. Cell. Mol. Life Sci. 2005, 62, 2576–2587. [Google Scholar] [CrossRef]
- Thomson, A.J.; Gray, H.B. Bio-inorganic chemistry. Curr. Opin. Chem. Biol. 1998, 2, 155–158. [Google Scholar] [CrossRef]
- Pomastowski, P.; Sprynsky, M.; Žuvela, P.; Rafińska, K.; Milanowski, M.; Liu, J.J.; Yi, M.; Buszewski, B. Silver-Lactoferrin Nanocomplexes as a Potent Antimicrobial Agent. J. Am. Chem. Soc. 2016, 138, 7899–7909. [Google Scholar] [CrossRef]
- Railean-Plugaru, V.; Pomastowski, P.; Meller, K.; Złoch, M.; Katarzyna, R.; Boguslaw, B. Lactococcus lactis as a safe and inexpensive source of bioactive silver composites. Appl. Microbiol. Biotechnol. 2017, 101, 7141–7153. [Google Scholar]
- Verwey, E.J.W.; Overbeek, J.T.G. Theory of the stability of lyophobic colloids. J. Colloid Sci. 1955, 10, 224–225. [Google Scholar] [CrossRef] [Green Version]
- Hedberg, Y.S.; Dobryden, I.; Chaudhary, H.; Wei, Z.; Claesson, P.M.; Lendel, C. Synergistic effects of metal-induced aggregation of human serum albumin. Colloids Surf. B Biointerfaces 2019, 173, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Li, D.; Chen, X.D.; Mercadé-Prieto, R. Effect of calcium on the fouling of whey protein isolate on stainless steel using QCM-D. Chem. Eng. Sci. 2018, 177, 501–508. [Google Scholar] [CrossRef]
- Magens, O.M.; Hofmans, J.F.A.; Adriaenssens, Y.; Ian Wilson, D. Comparison of fouling of raw milk and whey protein solution on stainless steel and fluorocarbon coated surfaces: Effects on fouling performance, deposit structure and composition. Chem. Eng. Sci. 2019, 195, 423–432. [Google Scholar] [CrossRef]
- Kluska, K.; Adamczyk, J.; Krężel, A. Metal binding properties, stability and reactivity of zinc fingers. Coord. Chem. Rev. 2018, 367, 18–64. [Google Scholar] [CrossRef]
- Garcia, J.S.; De Magalhães, C.S.; Arruda, M.A.Z. Trends in metal-binding and metalloprotein analysis. Talanta 2006, 69, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.; Au, K.; Francis, R.S.; Mudge, D.W.; Johnson, D.W.; Pillans, P.I. Phosphate binders in patients with chronic kidney disease. Aust. Prescr. 2017, 40, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, J.S.; Sadler, P.J. Targeted delivery of platinum-based anticancer complexes. Curr. Opin. Chem. Biol. 2013, 17, 175–188. [Google Scholar] [CrossRef] [Green Version]
- Henchion, M.; Hayes, M.; Mullen, A.; Fenelon, M.; Tiwari, B. Future Protein Supply and Demand: Strategies and Factors Influencing a Sustainable Equilibrium. Foods 2017, 6, 53. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, O.; Odani, A.; Takani, M. Metal–amino acid chemistry. Weak interactions and related functions of side chain groups. J. Chem. Soc. Dalt. Trans. 2002, 3411–3421. [Google Scholar] [CrossRef]
- Dudev, T.; Lim, C. Competition among metal ions for protein binding sites: Determinants of metal ion selectivity in proteins. Chem. Rev. 2014, 114, 538–556. [Google Scholar] [CrossRef]
- Tang, N.; Skibsted, L.H. Zinc bioavailability from whey. Enthalpy-entropy compensation in protein binding. Food Res. Int. 2016, 89, 749–755. [Google Scholar] [CrossRef]
- Udechukwu, M.C.; Downey, B.; Udenigwe, C.C. Influence of structural and surface properties of whey-derived peptides on zinc-chelating capacity, and in vitro gastric stability and bioaccessibility of the zinc-peptide complexes. Food Chem. 2018, 240, 1227–1232. [Google Scholar] [CrossRef]
- Shi, L.; Zhou, J.; Gunasekaran, S. Low temperature fabrication of ZnO-whey protein isolate nanocomposite. Mater. Lett. 2008, 62, 4383–4385. [Google Scholar] [CrossRef]
- Shahraki, S.; Shiri, F.; Majd, M.H.; Dahmardeh, S. Anti-cancer study and whey protein complexation of new lanthanum (III) complex with the aim of achieving bioactive anticancer metal-based drugs. J. Biomol. Struct. Dyn. 2018, 37, 2072–2085. [Google Scholar] [CrossRef] [PubMed]
- Shahraki, S.; Shiri, F.; Beyzaei, H.; Khosravi, F. Synthesis, characterization, protein interaction and antibacterial activity of a lanthanum(iii) complex [La(Trp)3(OH2)2] (Trp = tryptophan) as a new precursor for synthesis of La2O2CO3 nanoparticles. New J. Chem. 2017, 41, 8413–8421. [Google Scholar] [CrossRef]
- Shahraki, S.; Shiri, F.; Saeidifar, M. Evaluation of in silico ADMET analysis and human serum albumin interactions of a new lanthanum (III) complex by spectroscopic and molecular modeling studies. Inorganica Chim. Acta 2017, 463, 80–87. [Google Scholar] [CrossRef]
- Shahraki, S.; Heydari, A. Binding forces between a novel Schiff base palladium (II) complex and two carrier proteins: human serum albumi and β-lactoglobulin. J. Biomol. Struct. Dyn. 2018, 36, 2807–2821. [Google Scholar] [CrossRef]
- Zareian-Jahromi, S.; Mansouri-Torshizi, H. Synthesis, characterization, DNA and HSA binding studies of isomeric Pd (II) antitumor complexes using spectrophotometry techniques. J. Biomol. Struct. Dyn. 2018, 36, 1329–1350. [Google Scholar] [CrossRef] [PubMed]
- Shahraki, S.; Shiri, F.; Majd, M.H.; Razmara, Z. Comparative study on the anticancer activities and binding properties of a hetero metal binuclear complex [Co(dipic)2Ni(OH2)5]·2H2O (dipic = dipicolinate) with two carrier proteins. J. Pharm. Biomed. Anal. 2017, 145, 273–282. [Google Scholar] [CrossRef]
- Shahraki, S.; Shiri, F.; Razmara, Z.; Majd, M.H. A comparative study of the impact of metal complex size on the in vitro biological behavior of hetero di- and poly-nuclear Mn-Co complexes. J. Mol. Struct. 2019, 1178, 617–629. [Google Scholar] [CrossRef]
- Magyar, J.S.; Godwin, H.A. Spectropotentiometric analysis of metal binding to structural zinc-binding sites: Accounting quantitatively for pH and metal ion buffering effects. Anal. Biochem. 2003, 320, 39–54. [Google Scholar] [CrossRef]
- Kállay, C.; Várnagy, K.; Micera, G.; Sanna, D.; Sóvágó, I. Copper(II) complexes of oligopeptides containing aspartyl and glutamyl residues. Potentiometric and spectroscopic studies. J. Inorg. Biochem. 2005, 99, 1514–1525. [Google Scholar] [CrossRef]
- Casal, H.L.; Köhler, U.; Mantsch, H.H. Structural and conformational changes of β-lactoglobulin B: an infrared spectroscopic study of the effect of pH and temperature. Biochim. Biophys. Acta (BBA)/Protein Struct. Mol. 1988, 957, 11–20. [Google Scholar] [CrossRef]
- Herrero-Martínez, J.M.; Simó-Alfonso, E.F.; Ramis-Ramos, G.; Gelfi, C.; Righetti, P.G. Determination of cow’s milk in non-bovine and mixed cheeses by capillary electrophoresis of whey proteins in acidic isoelectric buffers. J. Chromatogr. A 2000, 878, 261–271. [Google Scholar] [CrossRef]
- Miralles, B.; Rothbauer, V.; Manso, M.A.; Amigo, L.; Krause, I.; Ramos, M. Improved method for the simultaneous determination of whey proteins, caseins and para-κ-casein in milk and dairy products by capillary electrophoresis. J. Chromatogr. A 2001, 915, 225–230. [Google Scholar] [CrossRef]
- Costa, F.F.; Vasconcelos Paiva Brito, M.A.; Moreira Furtado, M.A.; Martins, M.F.; Leal De Oliveira, M.A.; Mendonça De Castro Barra, P.; Amigo Garrido, L.; De Oliveira Dos Santos, A.S. Microfluidic chip electrophoresis investigation of major milk proteins: Study of buffer effects and quantitative approaching. Anal. Methods 2014, 6, 1666–1673. [Google Scholar] [CrossRef] [Green Version]
- Anema, S.G. The use of “lab-on-a-chip” microfluidic SDS electrophoresis technology for the separation and quantification of milk proteins. Int. Dairy J. 2009, 19, 198–204. [Google Scholar] [CrossRef]
- Buffoni, J.N.; Bonizzi, I.; Pauciullo, A.; Ramunno, L.; Feligini, M. Characterization of the major whey proteins from milk of Mediterranean water buffalo (Bubalus bubalis). Food Chem. 2011, 127, 1515–1520. [Google Scholar] [CrossRef]
- Li, Z.; Wen, F.; Li, Z.; Zheng, N.; Jiang, J.; Xu, D. Simultaneous detection of α-Lactoalbumin, β-Lactoglobulin and Lactoferrin in milk by Visualized Microarray. BMC Biotechnol. 2017, 17, 1–9. [Google Scholar] [CrossRef]
- Czerwenka, C.; Muller, L.; Lindner, W. Detection of the adulteration of water buffalo milk and mozzarella with cow’s milk by liquid chromatography-mass spectrometry analysis of β-lactoglobulin variants. Food Chem. 2010, 122, 901–908. [Google Scholar] [CrossRef]
- Santos, M.J.; Teixeira, J.A.; Rodrigues, L.R. Fractionation of the major whey proteins and isolation of β-Lactoglobulin variants by anion exchange chromatography. Sep. Purif. Technol. 2012, 90, 133–139. [Google Scholar] [CrossRef] [Green Version]
- Doultani, S.; Turhan, K.N.; Etzel, M.R. Fractionation of proteins from whey using cation exchange chromatography. Process Biochem. 2004, 39, 1737–1743. [Google Scholar] [CrossRef]
- Nicolás, P.; Ferreira, M.L.; Lassalle, V. A review of magnetic separation of whey proteins and potential application to whey proteins recovery, isolation and utilization. J. Food Eng. 2019, 246, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Barinov, N.A.; Vlasova, I.I.; Sokolov, A.V.; Kostevich, V.A.; Dubrovin, E.V.; Klinov, D.V. High-resolution atomic force microscopy visualization of metalloproteins and their complexes. Biochim. Biophys. Acta - Gen. Subj. 2018, 1862, 2862–2868. [Google Scholar] [CrossRef]
- Acosta, M.; Sabier, T.; Mariño-Repizo, L.; Martinez, L.D.; Gil, R.A. Novel method for metalloproteins determination in human breast milk by size exclusion chromatography coupled to inductively coupled plasma mass spectrometry. J. Pharm. Biomed. Anal. 2018, 158, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Kondapi, A.K. Receptor-mediated targeted delivery of DNA using Lactoferrin nanoparticles. Int. J. Biol. Macromol. 2018, 108, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Srisod, S.; Motina, K.; Inprasit, T.; Pisitsak, P. A green and facile approach to durable antimicrobial coating of cotton with silver nanoparticles, whey protein, and natural tannin. Prog. Org. Coatings 2018, 120, 123–131. [Google Scholar] [CrossRef]
- Kumari, S.; Kondapi, A.K. Lactoferrin nanoparticle mediated targeted delivery of 5-fluorouracil for enhanced therapeutic efficacy. Int. J. Biol. Macromol. 2017, 95, 232–237. [Google Scholar] [CrossRef]
- Bollimpelli, V.S.; Kumar, P.; Kumari, S.; Kondapi, A.K. Neuroprotective effect of curcumin-loaded lactoferrin nano particles against rotenone induced neurotoxicity. Neurochem. Int. 2016, 95, 37–45. [Google Scholar] [CrossRef]
- Okubo, K.; Kamiya, M.; Urano, Y.; Nishi, H.; Herter, J.M.; Mayadas, T.; Hirohama, D.; Suzuki, K.; Kawakami, H.; Tanaka, M.; et al. Lactoferrin Suppresses Neutrophil Extracellular Traps Release in Inflammation. EBioMedicine 2016, 10, 204–215. [Google Scholar] [CrossRef] [Green Version]
- Pozzi, C.M.C.; Braga, C.P.; Vieira, J.C.S.; Cavecci, B.; Vitor de Queiroz, J.; de Souza Barbosa, H.; Arruda, M.A.Z.; Gozzo, F.C.; Padillha, P.; de Magalhães Padilha, M. Metal ions bound to the human milk immunoglobulin A: Metalloproteomic approach. Food Chem. 2015, 166, 492–497. [Google Scholar] [CrossRef]
- Kumar, P.; Lakshmi, Y.S.; Bhaskar, C.; Golla, K.; Kondapi, A.K. Improved Safety, Bioavailability and Pharmacokinetics of Zidovudine through Lactoferrin Nanoparticles during Oral Administration in Rats. PLoS One 2015, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Xing, L.; Chen, Y.; Xu, Y.; Yang, F.; Zhang, C.; Ping, Q.; Xiao, Y. Lactoferrin-Modified Poly(ethylene glycol)-Grafted BSA Nanoparticles as a Dual-Targeting Carrier for Treating Brain Gliomas. Mol. Pharm. 2014, 11, 1823–1834. [Google Scholar] [CrossRef]
- Godoy-Gallardo, M.; Mas-Moruno, C.; Fernández-Calderón, M.C.; Pérez-Giraldo, C.; Manero, J.M.; Albericio, F.; Gil, F.J.; Rodríguez, D. Covalent immobilization of hLf1-11 peptide on a titanium surface reduces bacterial adhesion and biofilm formation. Acta Biomater. 2014, 10, 3522–3534. [Google Scholar] [CrossRef] [PubMed]
- Carthagena, L.; Becquart, P.; Hocini, H.; Kazatchkine, M.D.; Bouhlal, H.; Belec, L. Modulation of HIV Binding to Epithelial Cells and HIV Transfer from Immature Dendritic Cells to CD4 T Lymphocytes by Human Lactoferrin and its Major Exposed LF-33 Peptide. Open Virol. JournalOpen Virol. J. 2011, 5, 27–34. [Google Scholar] [CrossRef]
- Hutchens, T.W.; Magnuson, J.S.; Yip, T. Rapid purification of porcine colostral whey lactoferrin by affinity chromatography on single-stranded DNA-agarose. Characterization, amino acid composition and N-terminal amino acid sequence. Biochim. Biophys. Acta 1989, 999, 323–329. [Google Scholar] [CrossRef]
- Pomastowski, P.; Buszewski, B. Complementarity of matrix-and nanostructure-assisted laser desorption/ionization approaches. Nanomaterials 2019, 9, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leney, A.C.; Heck, A.J.R. Native Mass Spectrometry: What is in the Name? J. Am. Soc. Mass Spectrom. 2017, 28, 5–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lermyte; Everett; Brooks; Bellingeri; Billimoria; Sadler; O’Connor; Telling; Collingwood Emerging Approaches to Investigate the Influence of Transition Metals in the Proteinopathies. Cells 2019, 8, 1231. [CrossRef] [PubMed] [Green Version]
- Allen, S.J.; Giles, K.; Gilbert, T.; Bush, M.F. Ion mobility mass spectrometry of peptide, protein, and protein complex ions using a radio-frequency confining drift cell. Analyst 2016, 141, 884–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, S.J.; Schwartz, A.M.; Bush, M.F. Effects of polarity on the structures and charge states of native-like proteins and protein complexes in the gas phase. Anal. Chem. 2013, 85, 12055–12061. [Google Scholar] [CrossRef]
- Lermyte, F.; Everett, J.; Lam, Y.P.Y.; Wootton, C.A.; Brooks, J.; Barrow, M.P.; Telling, N.D.; Sadler, P.J.; O’Connor, P.B.; Collingwood, J.F. Metal Ion Binding to the Amyloid β Monomer Studied by Native Top-Down FTICR Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2019, 30, 2123–2134. [Google Scholar] [CrossRef]
- Lee, C.W.; Tseng, F.G. Surface enhanced Raman scattering (SERS) based biomicrofluidics systems for trace protein analysis. Biomicrofluidics 2018, 12. [Google Scholar] [CrossRef]
- Van De Weert, M.; Stella, L. Fluorescence quenching and ligand binding: A critical discussion of a popular methodology. J. Mol. Struct. 2011, 998, 144–150. [Google Scholar] [CrossRef]
- Kastritis, P.L.; Bonvin, A.M.J.J. On the binding affinity of macromolecular interactions: daring to ask why proteins interact. J. R. Soc. Interface 2012, 10, 20120835. [Google Scholar] [CrossRef] [PubMed]
- Vuignier, K.; Schappler, J.; Veuthey, J.L.; Carrupt, P.A.; Martel, S. Drug-protein binding: A critical review of analytical tools. Anal. Bioanal. Chem. 2010, 398, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Tai, C.S.; Chen, Y.Y.; Chen, W.L. β-Lactoglobulin Influences Human Immunity and Promotes Cell Proliferation. Biomed Res. Int. 2016, 2016, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Žuvela, P.; Liu, J.J.; Yi, M.; Pomastowski, P.P.; Sagandykova, G.; Belka, M.; David, J.; Bączek, T.; Szafrański, K.; Żołnowska, B.; et al. Target-based drug discovery through inversion of quantitative structure-drug-property relationships and molecular simulation: CA IX-sulphonamide complexes. J. Enzyme Inhib. Med. Chem. 2018, 33, 1430–1443. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Sun, L.; Yu, Y.; Xue, Y.; Yang, P. Amino acid-coded tagging approaches in quantitative proteomics. Expert Rev. Proteomics 2007, 4, 25–37. [Google Scholar] [CrossRef]
- Ross, P.L.; Huang, Y.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 2004, 3, 1154–1169. [Google Scholar] [CrossRef] [Green Version]
- Shenar, N.; Cantel, S.; Martinez, J.; Enjalbal, C. Comparison of inert supports in laser desorption/ionization mass spectrometry of peptides: pencil lead, porous silica gel, DIOS-chip and NALDITM target. Rapid Commun. Mass Spectrom. 2009, 23, 2371–2379. [Google Scholar] [CrossRef]
- Zarogoulidis, P.; Tsakiridis, K.; Karapantzou, C.; Lampaki, S.; Kioumis, I.; Pitsiou, G.; Papaiwannou, A.; Hohenforst-Schmidt, W.; Huang, H.; Kesisis, G.; et al. Use of Proteins as Biomarkers and Their Role in Carcinogenesis. J. Cancer 2015, 6, 9–18. [Google Scholar] [CrossRef]
- Shelly, C.; Lu, M.D. Glutathione synthesis. Biochim. Biophys. Acta 2013, 1830, 3143–3153. [Google Scholar]
- Britigan, B.E.; Serody, J.S.; Cohen, M.S. The role of lactoferrin as an anti-inflammatory molecule. Adv. Exp. Med. Biol. 1994, 357, 143–156. [Google Scholar] [PubMed]
- Luzi, C.; Brisdelli, F.; Iorio, R.; Bozzi, A.; Carnicelli, V.; Di Giulio, A.; Lizzi, A.R. Apoptotic effects of bovine apo-lactoferrin on HeLa tumor cells. Cell Biochem. Funct. 2017, 35, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, J.A.; Kanwar, J.R.; Kanwar, R.K. Iron-free and iron-saturated bovine lactoferrin inhibit survivin expression and differentially modulate apoptosis in breast cancer. BMC Cancer 2015, 15, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanwar, J.R.; Palmano, K.P.; Sun, X.; Kanwar, R.K.; Gupta, R.; Haggarty, N.; Rowan, A.; Ram, S.; Krissansen, G.W. “Iron-saturated” lactoferrin is a potent natural adjuvant for augmenting cancer chemotherapy. Immunol. Cell Biol. 2008, 86, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, L.; Calvo, M.; Brock, J.H. Biological role of lactoferrin. Arch. Dis. Child. 1992, 67, 657–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komatsu, A.; Satoh, T.; Wakabayashi, H.; Ikeda, F. Effects of bovine lactoferrin to oral Candida albicans and Candida glabrata isolates recovered from the saliva in elderly people. Odontology 2015, 103, 50–55. [Google Scholar] [CrossRef]
- Leboffe, L.; Giansanti, F.; Antonini, G. Antifungal and Antiparasitic Activities of Lactoferrin. Antiinfect. Agents Med. Chem. 2009, 8, 114–127. [Google Scholar] [CrossRef]
- Thawari, A.G.; Rao, C.P. Peroxidase-like Catalytic Activity of Copper-Mediated Protein-Inorganic Hybrid Nanoflowers and Nanofibers of β-Lactoglobulin and α-Lactalbumin: Synthesis, Spectral Characterization, Microscopic Features, and Catalytic Activity. ACS Appl. Mater. Interfaces 2016, 8, 10392–10402. [Google Scholar] [CrossRef] [Green Version]
- Wegmüller, R.; Tay, F.; Zeder, C.; Brnić, M.; Hurrell, R.F. Zinc Absorption by Young Adults from Supplemental Zinc Citrate Is Comparable with That from Zinc Gluconate and Higher than from Zinc Oxide. J. Nutr. 2014, 144, 132–136. [Google Scholar] [CrossRef] [Green Version]
- Prasad, A.S. Discovery of human zinc deficiency: 50 years later. J. Trace Elem. Med. Biol. 2012, 26, 66–69. [Google Scholar] [CrossRef]
- Udechukwu, M.C.; Collins, S.A.; Udenigwe, C.C. Prospects of enhancing dietary zinc bioavailability with food-derived zinc-chelating peptides. Food Funct. 2016, 7, 4137–4144. [Google Scholar] [CrossRef] [PubMed]
- Pryshchepa, O.; Sagandykova, G.N.; Pomastowski, P.; Railean-Plugaru, V.; Król, A.; Rogowska, A.; Rodzik, A.; Sprynskyy, M.; Buszewski, B. A New Approach for Spontaneous Silver Ions Immobilization onto Casein. Int. J. Mol. Sci. 2019, 20, 3864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Li, B.; Wang, B.; Xie, N. Degradation and antioxidant activities of peptides and zinc-peptide complexes during in vitro gastrointestinal digestion. Food Chem. 2015, 173, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Jellesen, M.S.; Rasmussen, A.A.; Hilbert, L.R. A review of metal release in the food industry. Mater. Corros. 2006, 57, 387–393. [Google Scholar] [CrossRef]
- Atapour, M.; Wei, Z.; Chaudhary, H.; Lendel, C.; Odnevall Wallinder, I.; Hedberg, Y. Metal release from stainless steel 316L in whey protein - And simulated milk solutions under static and stirring conditions. Food Control 2019, 101, 163–172. [Google Scholar] [CrossRef]
- Atapour, M.; Odnevall Wallinder, I.; Hedberg, Y. Stainless steel in simulated milk and whey protein solutions – Influence of grade on corrosion and metal release. Electrochim. Acta 2020, 331, 135428. [Google Scholar] [CrossRef]
- Nicolás, P.; Ferreira, M.L.; Lassalle, V. Magnetic solid-phase extraction: A nanotechnological strategy for cheese whey protein recovery. J. Food Eng. 2019, 263, 380–387. [Google Scholar] [CrossRef]


| Protein | Mol. Weight (kDa) | Theoretical mol. Weight (kDa)* | PTM | Method of Isolation/Purification | Identification | Ref. |
|---|---|---|---|---|---|---|
| β-LG | 18 | 18.277 | - | standard of β-LG (protein content > 90%) | SDS-PAGE MALDI-TOF-MS | [5] |
| 18.5 | ||||||
| β-LG | 18.3 | 18.277 | monomeric and the dimeric forms at pH 7.4 glycated β-lactoglobulin | β-LG was dissolved in 9.1 mM glucose in water, and the pH was adjusted to 7 with 50 mM phosphate buffer | MALDI-TOF-MS | [16] |
| 36.6 | ||||||
| β-LG | 17.4 | 18.277 | - | anion-exchange chromatography (DEAE-Sepharose) | SDS-PAGE | [17] |
| β-LG | 19.9 | 18.277 | proteins appeared as strings of spots, indicating their different isoforms with different charges as a result of PTMs occurring prior to secretion | precipitation via ammonium sulphate fractionation | 2-DE | [18] |
| α-LA | 16.2 | 16.247 | MALDI-MS | |||
| α-LA | 14.1 | 16.247 | small mass differences ruled out PTMs, such as phosphorylation and glycosylation | precipitation by ammonium sulphate | MALDI-TOF-MS | [19] |
| SA | 67.7 (SA) | 69.367 | glycosylation of specific milk proteins was shown to vary during lactation; no potential N-glycosylation and O-linked glycans (SA), known N-linked glycoprotein (LTF) | 0.5 mL of raw milk was centrifuged at 4 °C for 30 min, fat and cellular layers were removed; residual lipids were removed by addition of three volumes (1.5 mL) of 2:1 chloroform/methanol, agitation, retaining of supernatant; protein was precipitated from supernatant with ethanol overnight at 4 °C, followed by centrifugation; precipitate was re-suspended in 50 mM ammonium bicarbonate buffer (pH 7.5); glycans were separated by SDS-PAGE and extracted for MALDI-MS analysis | MALDI-MS | [20] |
| 79.8 (LTF) | ||||||
| LTF | 69.0 (SA) | 78.056 | LC–MS/MS | |||
| 78.0 (LTF) | ||||||
| LTF | 80.002 | 78.056 | - | milk was defatted by centrifugation, and the pH was then adjusted to 4.6 using hydrochloric acid; precipitated casein was removed by centrifugation | RP-LC–MS/MS | [21] |
| Metal/Conc. | Compound/Conc. | Interaction | Analytical method | Ref. |
|---|---|---|---|---|
| Zn2+ | strong binding affinities: | ITC | [60] | |
| LTF | 2.7 × 105 M−1 | |||
| BSA | 2.3 × 105 M−1 | |||
| α-LA | 1.5 × 105 M−1 | |||
| β-LG | 1.5 × 105 M−1 | |||
| Zn2+(6.23 mM) | α-LA (63.9 µM) | two sets of independent binding sites for zinc (II) | ITC | [34] |
| two ions bind with the binding constant of 4.53 × 104 M−1 | fluorescence | |||
| four ions bind with the binding constant of 963 M−1 | CD | |||
| electrostatic interactions | DSC | |||
| Zn2+ | whey-derived peptides | zinc chelation | FT-IR | [61] |
| electrostatic interactions | zinc chelating capacity | |||
| ZnO | WPI | DSC curves allowed to suggest; hydrogen bonding; O–Zn–O bonding; or electrostatic interactions; XRD and UV-Vis allowed to observe evidence for phase structure and crystal quality of ZnO nanoparticles; TEM—image of ZnO-WPI nanocomposite | XRD, TEM, DSC, UV-Vis | [62] |
| Ag+ | LTF | two stages: (i) internal diffusion and sorption onto the external surface of lactoferrin globules; (ii) internal diffusion and binding to the lactoferrin structure; via electrostatic and hydrophobic interactions | MALDI-TOF/TOF-MS, ICP-MS, FT-IR, SERS, TEM, EDX, electrophoretic techniques | [47] |
| La (III)-Cys complex | hydrogen bonds, van der Waals interactions | NMR, UV-Vis, FT-IR, TG-DTA, FRET, CD | [63] | |
| BSA | KBSA-La 0.11 × 104 M−1; | |||
| β-LG | Kβ-LG-La 0.63 × 103 M−1 | |||
| La (III)-Trp complex | hydrophobic interactions: | NMR, UV-Vis, FT-IR, TG-DTA | [64] | |
| HSA | Kb 0.138 × 104 M−1 (303 K) | |||
| La (III)-Phe complex | hydrogen bonds, hydrophobic interactions Kb 0.174 × 104 M−1 (303 K) | NMR, UV-Vis, FT-IR | [65] | |
| HSA | ||||
| Pd (II) complex | hydrogen bonds, van der Waals interactions | NMR, UV-Vis, FT-IR | [66] | |
| HSA (1 × 105 M) | Kb 0.5 × 104 M−1; | |||
| β-LG (1 × 105 M) | Kb 0.2 × 103 M−1 | |||
| Pd (II) complexes (10−4 M) | hydrogen bonds, van der Waals interactions | NMR, UV-Vis, FT-IR, FRET | [67] | |
| HSA (2 mg/mL) | I complex: Kb 0.49 × 104 M−1 (293 K); | |||
| II complex: Kb 0.79 × 104 M−1 (293 K) | ||||
| Co (II)-Ni (II) complexes | hydrogen bonds, van der Waals interactions | UV-Vis, FT-IR, fluorescence | [68] | |
| HSA | Kb 3.16 × 106 M−1 (303 K); | |||
| β-LG | Kb 0.54 × 105 M−1 (303 K) | |||
| Mn (II)-Co (II) complexes (5 × 10−3 M) | hydrogen bonds, hydrophobic interactions | UV-Vis, FT-IR, FRET | [69] | |
| HSA (5 × 10−4 M) | I: Kb 7.4 ± 0.04 × 104 M−1 (303 K); | |||
| II: Kb 6.08 ± 0.09 × 103 M−1 (303 K) | ||||
| β-LG (5 × 10−4 M) | I: Kb 7.13 ± 0.03 × 104 M−1 (303 K); | |||
| II: Kb 2.62 ± 0.05 × 103 M−1 (303 K) |
| Proteins | Matrix | Isolation | Separation | Identification | Ref. |
|---|---|---|---|---|---|
| α-LA | cheese | cheese extracts were desalted and preconcentrated using microcon membranes | CE with fused silica capillaries | DAD | [73] |
| β-LG A | |||||
| β-LG B | |||||
| β-LG | cow, goat, and ewe cheeses, incl. those of a single animal origin, binary ternary mixtures | desalted, preconcentrated samples were obtained with microcon membranes | CE with fused silica uncoated capillaries | DAD | [74] |
| α-LA | |||||
| α-LA | raw milk | mixture of standards of purified proteins, separation was achieved by adding SEP and TPS buffers to milk | SDS-PAGE; Microfluidic chip electrophoresis | Fluorescence | [75] |
| β-LG | |||||
| α-LA | fresh skim milk | mixed protein standards were prepared by combining each of the individual protein solutions (1 mL) | SDS-PAGE; Microfluidic chip electrophoresis | Fluorescence | [76] |
| β-LG | |||||
| caseins | |||||
| β-LG | milk | diluting 200 µL of ultracentrifuged whey with 400 µL of HPLC-grade water | LC, Jupiter C4 column; Microchip electrophoresis | UV, MS; Fluorescence | [77] |
| α-LA | |||||
| SA | |||||
| LTF | milk | samples were centrifuged to remove fat; skim milk was loaded onto lactoferrin immunoaffinity column | LC, Symmetry C4 Column | PDA | [78] |
| β-LG | |||||
| α-LA | |||||
| β-LG | buffalo mozzarella | mixtures of cow’s milk, water buffalo’s milk, mixtures of brine from cow’s milk mozzarella, brine from buffalo mozzarella were prepared in diff. vol. ratios for calibration purposes | LC, Supelco Discovery Bio Wide Pore C8 column | MS | [79] |
| α-LA | WPC | standard pure proteins to determine ret. times; equilibration buffer Tris-HCl at 20 mM; elution buffer Tris-HCl at 20 mM with 1 M NaCl were used for separation | Mono Q5/50 GL anion-exchange column, FPLC | UV-Vis; SDS-PAGE | [80] |
| β-LG | |||||
| BSA | |||||
| α-LA | mozzarella cheese whey | different equilibration and elution buffers were prepared | Chromatographic column; packed with SP Sepharose Big; Beads cation exchanger, HPLC | UV-Vis; SDS-PAGE | [81] |
| β-LG | |||||
| BSA |
| Compounds | Form | Application | Analytical methods | Ref. |
|---|---|---|---|---|
| LTF | metalloprotein | regulation of inflammation and oxidative stress in vertebrates | AFM | [83] |
| α-LA | metalloprotein | nutrition of infants in a long breastfeeding stage | Native-PAGE; SEC-ICP-MS; MALDI-TOF/TOF-MS | [84] |
| LF | ||||
| serum albumin | ||||
| LTF | nanoparticles | gene delivery carrier with targeting abilities | TEM | [85] |
| WPI | nanoparticles | production of antimicrobial cotton fabrics | UV-Vis; TEM; SEM | [86] |
| LTF | nanoparticles | increased therapeutic efficacy of treatment of malignant melanoma | TEM; SEM; DLS; FT-IR | [87] |
| LTF | metallocomposites; nanoparticles | in medicine and food industry as an antimicrobial agent | MALDI-TOF/TOF-MS; ICP-MS; FT-IR; SERS; TEM; I-DE; zeta potential measurements | [47] |
| LTF | nanoparticles | drug delivery strategy against the neurotoxicity in dopaminergic neurons | FE-SEM; AFM; DLS | [88] |
| LTF | metalloprotein | a therapeutic lead for controlling neutrophil extracellular traps (NETs) release in autoimmune and inflammatory diseases | TEM; SEM; fluorescence microscopy; agarose gel electrophoresis | [89] |
| IgA | metalloprotein | health and nutrition of breastfed newborns | ESI-MS/MS; FAAS | [90] |
| LTF | nanoparticles | target specific drug delivery, encapsulation of the drug | FE-SEM; AFM; FT-IR | [91] |
| LTF | nanoparticles | drug delivery for effective targeting therapy of brain glioma | Particle electrophoresis | [92] |
| BSA | ||||
| LTF | metalloprotein | antimicrobial biomaterials for dental applications | HPLC; SEM; XPS | [93] |
| LTF | metalloprotein | inhibition of the attachment of free HIV-1 to epithelial cells | ELISA; flow cytometry | [94] |
| LTF | Metalloprotein; metallocomplex | immobilized DNA effective for LTF purification | HPCEC; HPIMAC; HPLC; HPSEC; SDS-PAGE | [95] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodzik, A.; Pomastowski, P.; Sagandykova, G.N.; Buszewski, B. Interactions of Whey Proteins with Metal Ions. Int. J. Mol. Sci. 2020, 21, 2156. https://doi.org/10.3390/ijms21062156
Rodzik A, Pomastowski P, Sagandykova GN, Buszewski B. Interactions of Whey Proteins with Metal Ions. International Journal of Molecular Sciences. 2020; 21(6):2156. https://doi.org/10.3390/ijms21062156
Chicago/Turabian StyleRodzik, Agnieszka, Paweł Pomastowski, Gulyaim N. Sagandykova, and Bogusław Buszewski. 2020. "Interactions of Whey Proteins with Metal Ions" International Journal of Molecular Sciences 21, no. 6: 2156. https://doi.org/10.3390/ijms21062156
APA StyleRodzik, A., Pomastowski, P., Sagandykova, G. N., & Buszewski, B. (2020). Interactions of Whey Proteins with Metal Ions. International Journal of Molecular Sciences, 21(6), 2156. https://doi.org/10.3390/ijms21062156