Comparative Transcriptome Analysis Reveals the Potential Mechanism of Abortion in Tobacco sua-Cytoplasmic Male Sterility
Abstract
:1. Introduction
2. Results
2.1. Cytological Characteristics of Early Anther Development of msZY
2.2. RNA Sequencing and Identification of Differentially Expressed Genes (DEGs)
2.3. Gene Ontology (GO) Enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis of DEGs
2.4. Verification of Differentially Expressed Genes by qRT-PCR
2.5. Genes Related to Stamen Development
2.6. Differentially Expressed Genes Related to Energy Metabolism
2.7. Differentially Expressed Genes in Endoplasmic Reticulum Stress and Programmed Cell Death
2.8. Differentially Expressed Genes Related to the Heat Shock Protein Family
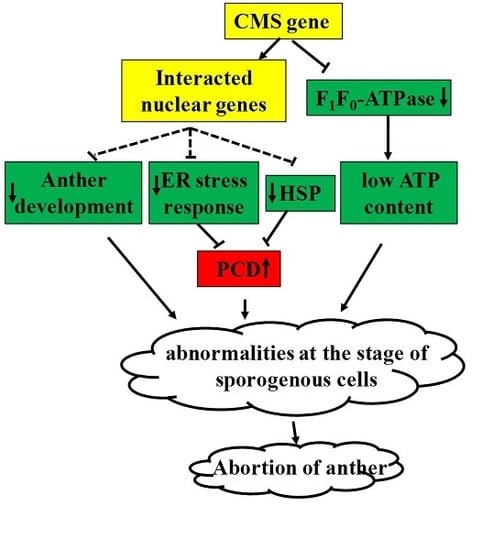
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Microscopic Observation of Early Anther Development
4.3. RNA-Seq
4.4. Sequence Assembly and Differentially Expressed Gene (DEG) Analysis
4.5. qRT-PCR Analysis
4.6. Quantification of ATP
4.7. Transmission Electron Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ER | endoplasmic reticulum |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PCD | programmed cell death |
| CMS | cytoplasmic male sterility |
| TFs | transcription factors |
| AP3 | APETALA3 |
| PI | PISTILLATA |
| AG | AGAMOUS |
| NZZ/SPL | NOZZLE/SPOROCYTELESS |
| OXPHOS | oxidative phosphorylation |
| ROS | reactive oxygen species |
| TCA cycle | tricarboxylic acid cycle |
| ERQC | ER quality control |
| UPR | unfolded protein response |
| BiP | binding protein |
| CNX | calnexin |
| CRT | calreticulin |
| PDI | protein disulfide isomerase |
| IRE1 | inositol-requiring enzyme 1 |
| ERAD | ER-assisted degradation |
| NRP | N-rich protein |
| NAC | NAM, ATAF1, CUC2 |
| VPE | vacuolar processing enzyme |
| BI-1 | Bax inhibitor 1 |
| HSPs | heat-shock proteins |
| DEGs | differentially expressed genes |
| TEM | transmission electron microscopy |
| TM | tunicamycin |
| FPKM | fragments per kb per million reads |
References
- Bohra, A.; Jha, U.C.; Adhimoolam, P.; Bisht, D.; Singh, N.P. Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell Rep. 2016, 35, 967–993. [Google Scholar] [CrossRef]
- Horn, R.; Gupta, K.J.; Colombo, N. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 2014, 19, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Hanson, M.R.; Bentolila, S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 2004, 16, S154–S169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chase, C.D. Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 2007, 23, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Sanders, P.M.; Bui, A.Q.; Weterings, K.; McIntire, K.N.; Hsu, Y.; Lee, P.Y.; Truong, M.T.; Beals, T.P.; Goldberg, R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sexual Plant Reprod. 1999, 11, 297–322. [Google Scholar] [CrossRef]
- Yang, S.; Terachi, T.; Yamagishi, H. Inhibition of chalcone synthase expression in anthers of Raphanus sativus with Ogura male sterile cytoplasm. Ann. Bot. 2008, 102, 483–489. [Google Scholar] [CrossRef] [Green Version]
- An, H.; Yang, Z.; Yi, B.; Wen, J.; Shen, J.; Tu, J.; Ma, C.; Fu, T. Comparative transcript profiling of the fertile and sterile flower buds of pol CMS in B. napus. BMC Genomics 2014, 15, 258. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zou, Y.; Li, X.; Zhang, Q.; Chen, L.; Wu, H.; Su, D.; Chen, Y.; Guo, J.; Luo, D.; et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 2006, 18, 676–687. [Google Scholar] [CrossRef] [Green Version]
- Luo, D.; Xu, H.; Liu, Z.; Guo, J.; Li, H.; Chen, L.; Fang, C.; Zhang, Q.; Bai, M.; Yao, N.; et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 2013, 45, 573–577. [Google Scholar] [CrossRef]
- Wang, K.; Gao, F.; Ji, Y.; Liu, Y.; Dan, Z.; Yang, P.; Zhu, Y.; Li, S. ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in Honglian cytoplasmic male sterilerice. New Phytol. 2013, 198, 408–418. [Google Scholar] [CrossRef]
- Heng, S.; Liu, S.; Xia, C.; Tang, H.; Xie, F.; Fu, T.; Wan, Z. Morphological and genetic characterization of a new cytoplasmic male sterility system (oxa CMS) in stem mustard (Brassica juncea). Theor. Appl. Genet. 2018, 131, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Touzet, P.; Meyer, E.H. Cytoplasmic male sterility and mitochondrial metabolism in plants. Mitochondrion 2014, 19, 166–171. [Google Scholar] [CrossRef]
- Kofer, W.; Glimelius, K.; Bonnett, H.T. Modifications of mitochondrial DNA cause changes in floral development in homeotic-like mutants of tobacco. Plant Cell 1991, 3, 759–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murai, K.; Takumi, S.; Koga, H.; Ogihara, Y. Pistillody, homeotic transformation of stamens into pistil-like structures, caused by nuclear-cytoplasm interaction in wheat. Plant J. 2002, 29, 169–181. [Google Scholar] [CrossRef]
- Heng, S.; Chen, F.; Wei, C.; Li, X.; Yi, B.; Ma, C.; Tu, J.; Shen, J.; Fu, T.; Wen, J. Cytological and iTRAQ-based quantitative proteomic analyses of hau CMS in Brassica napus L. J. Protemics 2019, 193, 230–238. [Google Scholar] [CrossRef] [PubMed]
- O’Maoileidigh, D.S.; Graciet, E.; Wellmer, F. Gene networks controlling Arabidopsis thaliana flower development. New Phytol. 2014, 201, 16–30. [Google Scholar] [CrossRef] [Green Version]
- Schiefthaler, U.; Balasubramanian, S.; Sieber, P.; Chevalier, D.; Wisman, E.; Schneitz, K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1999, 96, 11664–11669. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.Z.; Wang, G.F.; Speal, B.; Ma, H. The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Gene. Dev. 2002, 16, 2021–2031. [Google Scholar] [CrossRef] [Green Version]
- Wuest, S.E.; O’Maoileidigh, D.S.; Rae, L.; Kwasniewska, K.; Raganelli, A.; Hanczaryk, K.; Lohan, A.J.; Loftus, B.; Graciet, E.; Wellmer, F. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 2012, 109, 13452–13457. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Ye, X.; Zhang, S.; Zhu, S.; Yuan, L.; Hou, J.; Wang, C. Comparative transcriptome analysis between fertile and CMS flower buds in Wucai (Brassica campestris L.). BMC Genomics 2018, 19, 908. [Google Scholar] [CrossRef]
- Ning, L.; Wang, H.; Li, D.; Lin, Z.; Li, Y.; Zhao, W.; Chao, H.; Miao, L.; Li, M. Transcriptomic and proteomic analysis of Shaan2A cytoplasmic male sterility and its maintainer line in Brassica napus. Front. Plant Sci. 2019, 10, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Liu, Y. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef] [PubMed]
- Geisler, D.A.; Päpke, C.; Obata, T.; Nunes-Nesi, A.; Matthes, A.; Schneitz, K.; Maximova, E.; Araújo, W.L.; Fernie, A.R.; Persson, S. Downregulation of the δ-Subunit reduces mitochondrial ATP synthase levels, alters respiration, and restricts growth and gametophyte Development in Arabidopsis. Plant Cell 2012, 24, 2792–2811. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Pandeya, D.; Jo, Y.D.; Liu, W.Y.; Kang, B.C. Reduced activity of ATP synthase in mitochondria causes cytoplasmic male sterility in chili pepper. Planta 2013, 237, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Sabar, M.; Gagliardi, D.; Balk, J.; Leaver, C.J. ORFB is a subunit of F1F(O)-ATP synthase: Insight into the basis of cytoplasmic male sterility in sunflower. EMBO Rep. 2003, 4, 381–386. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, A.; Mitra, J.; Bhattacharyya, J.; Pradhan, S.; Sikdar, N.; Das, S.; Chakraborty, S.; Kumar, S.; Lakhanpaul, S.; Sen, S.K. Transgenic expression of an unedited mitochondrial orfB gene product from wild abortive (WA) cytoplasm of rice (Oryza sativa L.) generates male sterility in fertile rice lines. Planta 2015, 241, 1463–1479. [Google Scholar] [CrossRef]
- Zabaleta, E.; Mouras, A.; Hernould, M.; Suharsono; Araya, A. Transgenic male-sterile plant induced by an unedited atp9 gene is restored to fertility by inhibiting its expression with antisense RNA. Proc. Natl. Acad. Sci. USA 1996, 93, 11259–11263. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Srivastava, R.; Howell, S.H. Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int. J. Mol. Sci. 2013, 14, 8188–8212. [Google Scholar] [CrossRef] [Green Version]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef] [Green Version]
- Strasser, R. Protein quality control in the endoplasmic reticulum of plants. Annu. Rev. Plant Biol. 2018, 69, 147–172. [Google Scholar] [CrossRef]
- Iwata, Y.; Koizumi, N. Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci. 2012, 17, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Shi, G.; Sha, H.; Ji, Y.; Han, X.; Shu, X.; Ma, H.; Inoue, T.; Gao, B.; Kim, H.; et al. IRE1alpha is an endogenous substrate of endoplasmic-reticulum-associated degradation. Nat. Cell Biol. 2015, 17, 1546–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagashima, Y.; Mishiba, K.; Suzuki, E.; Shimada, Y.; Iwata, Y.; Koizumi, N. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 2011, 1, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, A.A.; Orellana, A. The physiological role of the unfolded protein response in plants. Biol. Res. 2011, 44, 75–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Howell, S.H. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 2010, 22, 2930–2942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Burgos, J.S.; Deng, Y.; Srivastava, R.; Howell, S.H.; Bassham, D.C. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 2012, 24, 4635–4651. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Wang, J.; Liu, Y.; Miao, H.; Cai, C.; Shao, Z.; Guo, R.; Sun, B.; Jia, C.; Zhang, L.; et al. Classic myrosinase-dependent degradation of indole glucosinolate attenuates fumonisin B1-induced programmed cell death in Arabidopsis. Plant J. 2015, 81, 920–933. [Google Scholar] [CrossRef]
- Daneva, A.; Gao, Z.; Van Durme, M.; Nowack, M.K. Functions and regulation of programmed cell death in plant development. Annu. Rev. Cell. Dev. Biol. 2016, 32, 441–468. [Google Scholar] [CrossRef]
- Balk, J.; Leaver, C.J. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 2001, 13, 1803–1818. [Google Scholar] [CrossRef] [Green Version]
- Haddad, I.; de Sa-Haiad, B.; de Santiago-Fernandes, L.; Machado, S.R. Pollen grain development and male sterility in the perfect flowers of Maytenusobtusifolia Mart. (Celastraceae). Protoplasma 2018, 256, 745–761. [Google Scholar] [CrossRef]
- Nugent, J.M.; Byrne, T.; McCormack, G.; Quiwa, M.; Stafford, E. Progressive programmed cell death inwards across the anther wall in male sterile flowers of the gynodioecious plant Plantagolanceolata. Planta 2019, 249, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Reis, P.A.; Carpinetti, P.A.; Freitas, P.P.; Santos, E.G.; Camargos, L.F.; Oliveira, I.H.; Silva, J.C.; Carvalho, H.H.; Dal-Bianco, M.; Soares-Ramos, J.R.; et al. Functional and regulatory conservation of the soybean ER stress-induced DCD/NRP-mediated cell death signaling in plants. BMC Plant Biol. 2016, 16, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendes, G.C.; Reis, P.A.; Calil, I.P.; Carvalho, H.H.; Aragao, F.J.; Fontes, E.P. GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc. Natl. Acad. Sci. USA 2013, 110, 19627–19632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatsugai, N.; Kuroyanagi, M.; Yamada, K.; Meshi, T.; Tsuda, S.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 2004, 305, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Lam, E. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J. Biol. Chem. 2008, 283, 3200–3210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Reed, J.C. Bax Inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol. Cell 1998, 1, 337–346. [Google Scholar] [CrossRef]
- McClellan, A.J.; Xia, Y.; Deutschbauer, A.M.; Davis, R.W.; Gerstein, M.; Frydman, J. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell 2007, 131, 121–135. [Google Scholar] [CrossRef] [Green Version]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Ito, M.; Yamamoto, Y.; Kim, C.S.; Ohnishi, K.; Hikichi, Y.; Kiba, A. Heat shock protein 70 is required for tabtoxinine-beta-lactam-induced cell death in Nicotiana benthamiana. J. Plant Physiol. 2014, 171, 173–178. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, H.; Zou, Y.; Liu, C.; Liu, Y.; Wang, Y.; Zhang, W. Over-expression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice. FEBS Lett. 2011, 585, 231–239. [Google Scholar] [CrossRef] [Green Version]
- Dickman, M.; Williams, B.; Li, Y.; de Figueiredo, P.; Wolpert, T. Reassessing apoptosis in plants. Nat. Plants 2017, 3, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, W.G.; Beers, E.P.; Dangl, J.L.; Franklin-Tong, V.E.; Gallois, P.; Hara-Nishimura, I.; Jones, A.M.; Kawai-Yamada, M.; Lam, E.; Mundy, J.; et al. Morphological classification of plant cell deaths. Cell Death Differ. 2011, 18, 1241–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatsugai, N.; Yamada, K.; Goto-Yamada, S.; Hara-Nishimura, I. Vacuolar processing enzyme in plant programmed cell death. Front. Plant Sci. 2015, 6, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, D. Tobacco Breeding; China Agricultural Press: Beijing, China, 1997. [Google Scholar]
- Zheng, Y.; Liu, Z.; Sun, Y.; Liu, G.; Yang, A.; Li, F. Characterization of genes specific to sua-CMS in Nicotiana tabacum. Plant Cell Rep. 2018, 37, 1245–1255. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, L.; Shi, Z.; Du, G.; Chen, J. ATP in current biotechnology: Regulation, applications and perspectives. Biotechnol. Adv. 2009, 27, 94–101. [Google Scholar] [CrossRef]
- Senkler, J.; Senkler, M.; Eubel, H.; Hildebrandt, T.; Lengwenus, C.; Schertl, P.; Schwarzlander, M.; Wagner, S.; Wittig, I.; Braun, H.P. The mitochondrial complexome of Arabidopsis thaliana. Plant J. 2017, 89, 1079–1092. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, Y.; Watase, Y.; Nagase, M.; Makita, N.; Yagura, S.; Hirai, A.; Sugiura, M. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: Comparative analysis of mitochondrial genomes in higher plants. Mol. Genet. Genomics 2005, 272, 603–615. [Google Scholar] [CrossRef]
- Du, K.; Xiao, Y.; Liu, Q.; Wu, X.; Jiang, J.; Wu, J.; Fang, Y.; Xiang, Y.; Wang, Y. Abnormal tapetum development and energy metabolism associated with sterility in SaNa-1A CMS of Brassica napus L. Plant Cell Rep. 2019, 38, 545–558. [Google Scholar] [CrossRef]
- Zhang, H.; Li, S.; Yi, P.; Wan, C.; Chen, Z.; Zhu, Y. A Honglian CMS line of rice displays aberrant F0 of F0F1-ATPase. Plant Cell Rep. 2007, 26, 1065–1071. [Google Scholar] [CrossRef]
- Wagner, S.; Steinbeck, J.; Fuchs, P.; Lichtenauer, S.; Elsässer, M.; Schippers, J.H.M.; Nietzel, T.; Ruberti, C.; Van Aken, O.; Meyer, A.J.; et al. Multiparametric real-time sensing of cytosolic physiology links hypoxia responses to mitochondrial electron transport. New Phytol. 2019, 224, 1668–1684. [Google Scholar] [CrossRef]
- Van der Linde, K.; Walbot, V. Pre-meiotic anther development. Curr. Top Dev. Biol. 2019, 131, 239–256. [Google Scholar] [PubMed]
- Amin-Wetzel, N.; Saunders, R.A.; Kamphuis, M.J.; Rato, C.; Preissler, S.; Harding, H.P.; Ron, D. A J-protein co-chaperone recruits BiP to monomerize IRE1 and repress the unfolded protein response. Cell 2017, 171, 1625–1637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leborgne-Castel, N.; Dooren, E.P.W.M.; Crofts, A.J.; Denecke, J. Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell 1999, 11, 459–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, P.A.; Rosado, G.L.; Silva, L.A.; Oliveira, L.C.; Oliveira, L.B.; Costa, M.D.; Alvim, F.C.; Fontes, E.P. The binding protein BiP attenuates stress-induced cell death in soybean via modulation of the N-rich protein-mediated signaling pathway. Plant Physiol. 2011, 157, 1853–1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishiba, K.; Nagashima, Y.; Suzuki, E.; Hayashi, N.; Ogata, Y.; Shimada, Y.; Koizumi, N. Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. USA 2013, 110, 5713–5718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.T.; Wang, M.J.; Sun, L.; Lu, S.J.; Bi, D.L.; Sun, L.; Song, Z.T.; Zhang, S.S.; Zhou, S.F.; Liu, J.X. The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 2014, 10, e1004243. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wan, C.; Kong, J.; Zhang, Z.; Li, Y.; Zhu, Y. Programmed cell death during microgenesis in a Honglian CMS line of rice is correlated with oxidative stress in mitochondria. Funct. Plant Biol. 2004, 31, 369. [Google Scholar] [CrossRef]
- Zuppini, A.; Navazio, L.; Mariani, P. Endoplasmic reticulum stress-induced programmed cell death in soybean cells. J. Cell Sci. 2004, 117, 2591–2598. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.M.; Yu, J.; Gallois, P. Endoplasmic reticulum stress-induced PCD and caspase-like activities involved. Front. Plant Sci. 2014, 5, 41. [Google Scholar] [CrossRef] [Green Version]
- Cronje, M.J.; Weir, I.E.; Bornman, L. Salicylic acid-mediated potentiation of Hsp70 induction correlates with reduced apoptosis in tobacco protoplasts. Cytometry Part A 2004, 61, 76–87. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatis 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]






| Pathway of Function | Gene | Gene ID | msZY-FPKM | ZY-FPKM | Fold Change (msZY/ZY) | p-Value |
|---|---|---|---|---|---|---|
| ER stress | BiP | LOC107771425 | 27.5 | 64.4 | 0.4 | 1.2 × 10−7 |
| bZIP60 | LOC107761889 | 14.2 | 49.1 | 0.3 | 0.00014685 | |
| CNX | LOC107772248 | 62.7 | 81.4 | 0.8 | 0.041817 | |
| CRT | LOC107814655 | 156.2 | 197.9 | 0.8 | 2.60 × 10−81 | |
| HRD1 | LOC107832607 | 1.1 | 1.4 | 0.8 | 1.25 × 10−76 | |
| HRD1 | LOC107781322 | 7.9 | 9.4 | 0.8 | 8.35 × 10−75 | |
| HRD3A | LOC107799383 | 6.5 | 8.7 | 0.7 | 1.03 × 10−74 | |
| PCD | ERD15 | LOC107831960 | 58.5 | 12.4 | 4.7 | 0.00000922 |
| NAC81 | LOC107784516 | 0.6 | 0.4 | 1.6 | 0.0058003 | |
| NRP-A/B | LOC107799405 | 200.1 | 88.0 | 2.3 | 0.0 20581 | |
| NRP-A/B | LOC107786583 | 150.6 | 59.7 | 2.5 | 0.04181 | |
| VPE | LOC107807349 | 5.5 | 3.0 | 1.9 | 0.0016425 | |
| BI-1 | LOC107803860 | 0.6 | 1.1 | 0.5 | 0.046784 | |
| Anther development | SPL/NZZ | LOC107789918 | 0.21 | 5.50 | 0.04 | 1.25× 10−7 |
| PI | LOC107803851 | 25.48 | 51.21 | 0.50 | 0.0 20896 | |
| AG | LOC107761813 | 15.13 | 23.30 | 0.65 | 0.0022628 | |
| F1F0-ATPase | atp9 | - | 276.2 | 432.9 | 0.6 | 0.0012869 |
| ORFB(atp8) | - | 73.7 | 122.0 | 0.6 | 5.92 × 10−119 | |
| atp6 | - | 692.0 | 1104.6 | 0.6 | 2.35 × 10−260 | |
| atp4 | - | 146.9 | 212.0 | 0.7 | 1.75 × 10−16 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Liu, Y.; Sun, Y.; Yang, A.; Li, F. Comparative Transcriptome Analysis Reveals the Potential Mechanism of Abortion in Tobacco sua-Cytoplasmic Male Sterility. Int. J. Mol. Sci. 2020, 21, 2445. https://doi.org/10.3390/ijms21072445
Liu Z, Liu Y, Sun Y, Yang A, Li F. Comparative Transcriptome Analysis Reveals the Potential Mechanism of Abortion in Tobacco sua-Cytoplasmic Male Sterility. International Journal of Molecular Sciences. 2020; 21(7):2445. https://doi.org/10.3390/ijms21072445
Chicago/Turabian StyleLiu, Zhiwen, Yanfang Liu, Yuhe Sun, Aiguo Yang, and Fengxia Li. 2020. "Comparative Transcriptome Analysis Reveals the Potential Mechanism of Abortion in Tobacco sua-Cytoplasmic Male Sterility" International Journal of Molecular Sciences 21, no. 7: 2445. https://doi.org/10.3390/ijms21072445
APA StyleLiu, Z., Liu, Y., Sun, Y., Yang, A., & Li, F. (2020). Comparative Transcriptome Analysis Reveals the Potential Mechanism of Abortion in Tobacco sua-Cytoplasmic Male Sterility. International Journal of Molecular Sciences, 21(7), 2445. https://doi.org/10.3390/ijms21072445