A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction
Abstract
:1. Introduction
2. Results
2.1. hSNF5171–258 and BAF155SWIRM form a Heterodimer
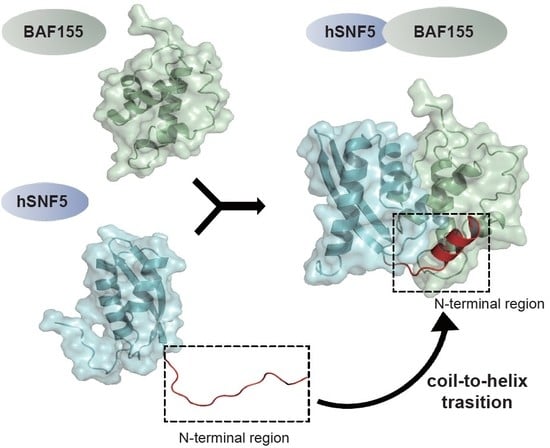
2.2. An N-Terminal Loop near the hSNF5RPT1 Domain Serves as a Binding Motif of BAF155SWIRM
2.3. N-Terminal Loop of hSNF5RPT1 Reveals Conformational Change upon BAF155SWIRM Binding
2.4. The Interface between hSNF5171–258 and BAF155SWIRM Features Charge Complementarity
2.5. The N-Terminal Binding Motif of hSNF5171–258 Enhances the Molecular Interaction for Stabilizing hSNF5171–258/BAF155SWIRM Complex
3. Discussion
4. Materials and Methods
4.1. Cloning, Protein Expression, and Purification
4.2. Size Exclusion Chromatography and Multi-Angle Light Scattering Analysis
4.3. NMR Spectroscopy
4.4. NMR Structure Determination and Analysis
4.5. Crystallization and Structure Determination
4.6. Crystallization and Structure Determination
4.7. Circular Dichroism Spectroscopy
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Accession codes
Abbreviations
| SWI/SNF | SWItch/Sucrose Non-Fermentable |
| RPT1 | repeat 1 |
| RPT2 | repeat2 |
| NMR | nuclear magnetic resonance |
| HSQC | heteronuclear single quantum coherence |
| CSP | chemical-shift perturbation |
| PDB | Protein Data Bank |
| NOEs | Overhauser effects |
| XNOE | 1H–15N heteronuclear NOE |
| ITC | isothermal titration calorimetry |
| IN | HIV-1 integrase |
| TEV | tobacco etch virus |
| IPTG | isopropyl-β-D-thiogalactopyranoside |
| TCEP | Tris(2-carboxyethyl)-phosphine |
| BMRB | Biological Magnetic Resonance Bank |
References
- Papamichos-Chronakis, E.; Peterson, C.L. Chromatin and the genome integrity network. Nat. Rev. Genet. 2013, 14, 62–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosgrove, M.S.; Boeke, J.D.; Wolberger, C. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 2004, 11, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Workman, J.L.; Kingston, R.E. Alteration of Nucleosome Structure As a Mechanism of Transcriptional Regulation. Annu. Rev. Biochem. 1998, 67, 545–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barkess, G. Chromatin remodeling and genome stability. Genome Biol. 2006, 7, 6–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, C.W.M.; Orkin, S.H. The SWI/SNF complex-Chromatin and cancer. Nat. Rev. Cancer 2004, 4, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Côté, J.; Xue, Y.; Zhou, S.; Khavari, P.A.; Biggar, S.R.; Muchardt, C.; Kalpana, G.V.; Goff, S.P.; Yaniv, M.; et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996, 15, 5370–5382. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, G.R. Diversity and specialization of mammalian SWI / SNF complexes. 2000, 4, 2117–2130. [Google Scholar]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef]
- Khavari, P.A.; Peterson, C.L.; Tamkun, J.W.; Mendel, D.B.; Crabtree, G.R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 1993, 366, 170–174. [Google Scholar] [CrossRef]
- Vignali, M.; Hassan, A.H.; Neely, K.E.; Workman, J.L. MINIREVIEW ATP-Dependent Chromatin-Remodeling Complexes. Mol. Cell. Biol. 2000, 20, 1899–1910. [Google Scholar] [CrossRef] [Green Version]
- Reisman, D.; Glaros, S.; Thompson, E.A. The SWI/SNF complex and cancer. Oncogene 2009, 28, 1653–1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.I. Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer Epigenetic Regulation of Transcription during Development Brg1 / Brm-Associated Factors (BAF), A Mammalian SWI / SNF-like ATP-dependent. Breast 2012, 44, 54–69. [Google Scholar]
- Kadoch, C.; Crabtree, G.R. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Rev. Clin. Esp. 2015, 197, 804–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clapier, C.R.; Cairns, B.R. The Biology of Chromatin Remodeling Complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Sarnowska, E.; Gratkowska, D.M.; Sacharowski, S.P.; Cwiek, P.; Tohge, T.; Fernie, A.R.; Siedlecki, J.A.; Koncz, C.; Sarnowski, T.J. The Role of SWI/SNF Chromatin Remodeling Complexes in Hormone Crosstalk. Trends Plant Sci. 2016, 21, 594–608. [Google Scholar] [CrossRef] [PubMed]
- Lessard, J.A.; Crabtree, G.R. Chromatin Regulatory Mechanisms in Pluripotency. Annu. Rev. Cell Dev. Biol. 2010, 26, 503–532. [Google Scholar] [CrossRef] [Green Version]
- Hughes, A.L.; Owen-Hughes, T. Deciphering Subunit-Specific Functions within SWI/SNF Complexes. Cell Rep. 2017, 18, 2075–2076. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.I.; Lessard, J.; Crabtree, G.R. Understanding the Words of Chromatin Regulation. Cell 2009. [Google Scholar] [CrossRef] [Green Version]
- Kalpana, G.V.; Marmon, S.; Wang, W.; Crabtree, G.R.; Goff, S.P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science (80-) 1994, 266, 2002–2006. [Google Scholar] [CrossRef]
- Morozov, A.; Yung, E.; Kalpana, G.V. Structure-function analysis of integrase interactor 1/hSNF5L1 reveals differential properties of two repeat motifs present in the highly conserved region. Proc. Natl. Acad. Sci. USA 1998, 95, 1120–1125. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.W.G.; Davies, K.P.; Yung, E.; Beltran, R.J.; Yu, J.; Kalpana, G.V. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet. 1999, 22, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Kim, J.W.; Seo, T.; Hwang, S.G.; Choi, E.J.; Choe, J. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 2002, 277, 22330–22337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maillot, B.; Lévy, N.; Eiler, S.; Crucifix, C.; Granger, F.; Richert, L.; Didier, P.; Godet, J.; Pradeau-Aubreton, K.; Emiliani, S.; et al. Structural and Functional Role of INI1 and LEDGF in the HIV-1 Preintegration Complex. PLoS ONE 2013, 8, e60734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, S.; Nguyen, M.; Wu, X.; Pal, A.; Shah, V.B.; Prasad, V.R.; Aiken, C.; Kalpana, G.V. INI1/hSNF5-interaction defective HIV-1 IN mutants exhibit impaired particle morphology, reverse transcription and integration in vivo. Retrovirology 2013, 10, 1. [Google Scholar] [CrossRef] [Green Version]
- Kohashi, K.; Oda, Y. Oncogenic roles of SMARCB1/INI1 and its deficient tumors. Cancer Sci. 2017, 108, 547–552. [Google Scholar] [CrossRef] [Green Version]
- Versteege, I.; Sévenet, N.; Lange, J.; Rousseau-Merck, M.F.; Ambros, P.; Handgretinger, R.; Aurias, A.; Delattre, O. Truncatingmutations of hSNF5/INI1 inaggressive paediatric cancer. 1998, 394, 203–206. [Google Scholar]
- Phelan, M.L.; Sif, S.; Narlikar, G.J.; Kingston, R.E. Reconstitution of a Core Chromatin Remodeling Complex from SWI/SNF Subunits. Mol. Cell 1999, 3, 247–253. [Google Scholar] [CrossRef]
- Alpsoy, A.; Dykhuizen, E.C. Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J. Biol. Chem. 2018, 293, 3892–3903. [Google Scholar] [CrossRef] [Green Version]
- Sohn, D.H.; Lee, K.Y.; Lee, C.; Oh, J.; Chung, H.; Jeon, S.H.; Seong, R.H. SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J. Biol. Chem. 2007, 282, 10614–10624. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.K.; Huh, S.-O.; Choi, H.; Lee, K.-S.; Shin, D.; Lee, C.; Nam, J.-S.; Kim, H.; Chung, H.; Lee, H.W.; et al. Srg3, a Mouse Homolog of Yeast SWI3, Is Essential for Early Embryogenesis and Involved in Brain Development. Mol. Cell. Biol. 2001, 21, 7787–7795. [Google Scholar] [CrossRef] [Green Version]
- Salbaum, J.M.; Niswander, L. A Unique Missense Allele of BAF155, a Core BAF Chromatin Remodeling Complex Protein, Causes Neural Tube Closure Defects in Mice. Dev. Neurobiol. 2015, 74, 483–497. [Google Scholar]
- Chen, J.; Archer, T.K. Regulating SWI / SNF Subunit Levels via Protein-Protein Interactions and Proteasomal Degradation: BAF155 and BAF170 Limit Expression of BAF57 Regulating SWI / SNF Subunit Levels via Protein-Protein Interactions and Proteasomal Degradation: BAF155 and BA. Mol. Cell. Biol. 2005, 25, 9016–9027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Bove, J.; Rosson, G.; Strobeck, M.; Chen, J.; Archer, T.K.; Wang, W.; Knudsen, E.S.; Weissman, B.E. Identification of a core member of the SWI/SNF complex, BAF155/SMARCC1, as a human tumor suppressor gene. Epigenetics 2011, 6, 1444–1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, L.; Xie, S.; Du, Y.; Qian, C. Structural Insights into BAF47 and BAF155 Complex Formation. J. Mol. Biol. 2017, 429, 1650–1660. [Google Scholar] [CrossRef] [PubMed]
- Sammak, S.; Allen, M.D.; Hamdani, N.; Bycroft, M.; Zinzalla, G. The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex. FEBS J. 2018, 285, 4165–4180. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef] [Green Version]
- Ruijtenberg, S.; van den Heuvel, S. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 2016, 15, 196–212. [Google Scholar] [CrossRef] [Green Version]
- Hohmann, A.F.; Vakoc, C.R. A rationale to target the SWI/SNF complex for cancer therapy. Trends Genet. 2014, 30, 356–363. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Reyes, A.A.; Malik, S.; He, Y. Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature 2020, 579, 452–455. [Google Scholar] [CrossRef]
- Pinto, E.M.; Hamideh, D.; Bahrami, A.; Orr, B.A.; Lin, T.; Pounds, S.; Zambetti, G.P.; Pappo, A.S.; Gajjar, A.; Agnihotri, S.; et al. Malignant rhabdoid tumors originating within and outside the central nervous system are clinically and molecularly heterogeneous. Acta Neuropathol. 2018, 136, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Uno, K.; Takita, J.; Yokomori, K.; Tanaka, Y.; Ohta, S.; Shimada, H.; Gilles, F.H.; Sugita, K.; Abe, S.; Sako, M.; et al. Aberrations of the hSNF5/INII gene are restricted to malignant rhabdoid tumors or atypical teratoid/rhabdoid tumors in pediatric solid tumors. Genes Chromosom. Cancer 2002, 34, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Cano, J.; Kalpana, G.V. Multimerization and DNA binding properties of INI1/hSNF5 and its functional significance. J. Biol. Chem. 2009, 284, 19903–19914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iakoucheva, L.M.; Brown, C.J.; Lawson, J.D.; Obradović, Z.; Dunker, A.K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002, 323, 573–584. [Google Scholar] [CrossRef] [Green Version]
- Galea, C.A.; Wang, Y.; Sivakolundu, S.G.; Kriwacki, R.W. Regulation of cell division by intrinsically unstructured proteins: Intrinsic flexibility, modularity, and signaling conduits. Biochemistry 2008, 47, 7598–7609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Perumal, N.B.; Oldfield, C.J.; Su, E.W.; Uversky, V.N.; Dunker, A.K. Intrinsic Disorder in Transcription Factors†. Biochem. 2006, 45, 6873–6888. [Google Scholar] [CrossRef] [Green Version]
- Sugase, K.; Dyson, H.J.; Wright, P.E. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 2007, 447, 1021–1025. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Bax, A. Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J. Biomol. NMR 2007, 38, 289–302. [Google Scholar] [CrossRef]
- Tjandra, N.; Wingfield, P.; Stahl, S.; Bax, A. Anisotropic rotational diffusion of perdeuterated HIV protease from 15N NMR relaxation measurements at two magnetic fields. J. Biomol. NMR 1996, 8, 273–284. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Tonelli, M.; Markley, J.L. Nmrfam-Sparky: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Güntert, P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar] [PubMed]
- Morris, A.L.; MacArthur, M.W.; Hutchinson, E.G.; Thornton, J.M. Stereochemical quality of protein structure coordinates. Proteins Struct. Funct. Bioinforma. 1992, 12, 345–364. [Google Scholar] [CrossRef] [PubMed]
- Koradi, R.; Billeter, M.; Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Vagin, A.; Teplyakov, A. MOLREP: An Automated Program for Molecular Replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Williams, J.C.; Zeelen, J.P.; Neubauer, G.; Vriend, G.; Backmann, J.; Michels, P.A.M.; Lambeir, A.M.; Wierenga, R.K. Structural and mutagenesis studies of leishmania triosephosphate isomerase: A point mutation can convert a mesophilic enzyme into a superstable enzyme without losing catalytic power. Protein Eng. 1999, 12, 243–250. [Google Scholar] [CrossRef] [Green Version]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2007, 1, 2527–2535. [Google Scholar] [CrossRef]






| Experimental Restraints | <SA> * |
|---|---|
| Non-redundant NOEs | 1891 |
| Dihedral angles, φ/ψ | 62/62 |
| Hydrogen bonds | 25 |
| Residual dipolar coupling, 1DNH | 77 |
| Total number of restraints | 1782 (24.3 per residue) |
| RMSD from experimental restraints | |
| Distances (Å) (1891) | 0.034 ± 0.002 |
| Torsion angles (°) (124) | 2.34 ± 0.14 |
| Residual dipolar coupling R-factor (%) † | |
| 1DNH (%) (77) | 2.9 ± 0.4 |
| RMSD from idealized covalent geometry | |
| Bonds (Å) | 0.003 ± 0 |
| Angles (°) | 0.59 ± 0.01 |
| Impropers (°) | 0.67 ± 0.02 |
| Coordinate precision (Å) *‡ | |
| Backbone | 0.48 ± 0.15 |
| Heavy atoms | 1.29 ± 0.17 |
| Ramachandran statistics (%) ठ| |
| Most favorable regions | 80.5 ± 1.1 |
| Allowed regions | 19.5 ± 1.1 |
| hSNF5171–258/BAF155SWIRM Complex | |
|---|---|
| Data Collection | |
| Space group | H3 |
| Cell dimensions | |
| a, b, c (Å) | 77.23, 77.23, 207.24 |
| α, β, γ (°) | 90.00, 90.00, 120.00 |
| Resolution (Å)a | 28.1–2.28 (2.236–2.28) |
| I/σI | 20.16 (3.01) |
| Rmerge (%) a | 8.5 (40.5) |
| Completeness (%) a | 99.87 (99.9) |
| Redundancy a | 5.43 (4.8) |
| Refinement | |
| Resolution (Å) | 28.1–2.28 (2.236–2.28) |
| No. reflections | 20987 |
| Rwork/Rfree | 0.1671/0.2003 |
| No. atoms | |
| Protein | 2876 |
| water | 223 |
| B-factors | |
| Protein | 33.21 |
| Water | 32.54 |
| RMSD | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.58 |
| Ramachandran plot (%) b | |
| Most favored regions | 98.55% |
| Allowed regions | 1.45% |
| Disallowed regions | 0% |
| Description | KD (nM) | ΔG (kcal/mol) | ΔH (kcal/mol) | −TΔS (kcal/mol) | |
|---|---|---|---|---|---|
| hSNF5 | BAF155SWIRM | ||||
| 171–253 a | Wild-type | 110 ± 20 | −9.5 ± 0.1 | −19.7 ± 0.3 | 10.2 ± 0.3 |
| 174–253 a | Wild-type | 110 ± 30 | −9.5 ± 0.2 | −15.8 ± 0.5 | 6.3 ± 0.3 |
| 179–253 a | Wild-type | 300 ± 50 | −8.9 ± 0.1 | −21.0 ± 0.6 | 12.1 ± 0.6 |
| 181–253 a | Wild-type | 300 ± 70 | −8.9 ± 0.1 | −12.9 ± 0.4 | 4.0 ± 0.5 |
| 183–253 a | Wild-type | 290 ± 80 | −8.9 ± 0.2 | −18.7 ± 0.8 | 9.8 ± 0.8 |
| 186–253 a | Wild-type | 760 ± 80 | −8.4 ± 0.1 | −16.0 ± 0.3 | 7.6 ± 0.3 |
| 171–253/E184Ab | Wild-type | 370 ± 100 | −8.8 ± 0.2 | −11.2 ± 0.4 | 2.4 ± 0.4 |
| 171–253 | N476A/N479Ad | 140 ± 30 | −9.3 ± 0.1 | −17.7 ± 0.7 | 8.3 ± 0.3 |
| SFH1/171–253c | Wild-type | 480 ± 140 | −8.6 ± 0.2 | −13.4 ± 0.6 | 4.8 ± 0.6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.; Kim, I.; Park, J.-H.; Yun, J.-H.; Joo, K.; Kim, T.; Park, G.-Y.; Ryu, K.-S.; Ko, Y.-J.; Mizutani, K.; et al. A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction. Int. J. Mol. Sci. 2020, 21, 2452. https://doi.org/10.3390/ijms21072452
Han J, Kim I, Park J-H, Yun J-H, Joo K, Kim T, Park G-Y, Ryu K-S, Ko Y-J, Mizutani K, et al. A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction. International Journal of Molecular Sciences. 2020; 21(7):2452. https://doi.org/10.3390/ijms21072452
Chicago/Turabian StyleHan, Jeongmin, Iktae Kim, Jae-Hyun Park, Ji-Hye Yun, Keehyoung Joo, Taehee Kim, Gye-Young Park, Kyoung-Seok Ryu, Yoon-Joo Ko, Kenji Mizutani, and et al. 2020. "A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction" International Journal of Molecular Sciences 21, no. 7: 2452. https://doi.org/10.3390/ijms21072452
APA StyleHan, J., Kim, I., Park, J. -H., Yun, J. -H., Joo, K., Kim, T., Park, G. -Y., Ryu, K. -S., Ko, Y. -J., Mizutani, K., Park, S. -Y., Seong, R. H., Lee, J., Suh, J. -Y., & Lee, W. (2020). A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction. International Journal of Molecular Sciences, 21(7), 2452. https://doi.org/10.3390/ijms21072452