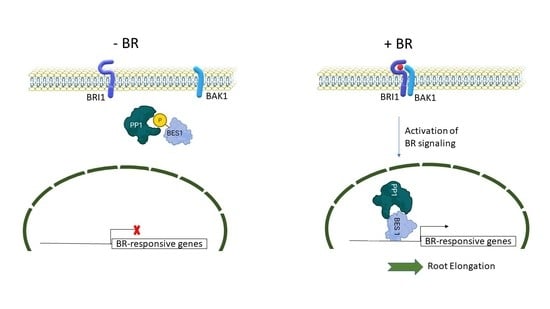
Wheat Type One Protein Phosphatase Participates in the Brassinosteroid Control of Root Growth via Activation of BES1
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Transgenic Lines
4.3. eBL Treatment and Meristem Size Measurement
4.4. Phosphorylation Status of BES1
4.5. Western Blotting
4.6. Protein Extraction and Co-Immunoprecipitation (co-IP) Assays
4.7. Bimolecular Fluorescence Complementation (BiFC) Assays
4.8. Real-Time Quantitative RT-PCR (qRT-PCR)
4.9. Confocal Imaging of Arabidopsis and N. Benthamiana
4.10. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Belkhadir, Y.; Jaillais, Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015, 206, 522–540. [Google Scholar] [CrossRef] [PubMed]
- Sharma, I.; Kaur, N.; Pati, P.K. Brassinosteroids: A Promising Option in Deciphering Remedial Strategies for Abiotic Stress Tolerance in Rice. Front. Plant Sci. 2017, 8, 2151. [Google Scholar] [CrossRef]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383. [Google Scholar] [CrossRef]
- Nam, K.H.; Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.W.; Zhou, H.W.; Deng, Z.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukasinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Liu, M.; Yuan, M.; Oses-Prieto, J.A.; Cai, X.; Sun, Y.; Burlingame, A.L.; Wang, Z.-Y.Z.Y.; Tang, W. The Brassinosteroid-Activated BRI1 Receptor Kinase Is Switched off by Dephosphorylation Mediated by Cytoplasm-Localized PP2A B′ Subunits. Mol. Plant 2016, 9, 148–157. [Google Scholar] [CrossRef]
- Farkas, I.; Dombrádi, V.; Miskei, M.; Szabados, L.; Koncz, C. Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 2007, 12, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Uhrig, R.G.; Labandera, A.M.; Moorhead, G.B. Arabidopsis PPP family of serine/threonine protein phosphatases: Many targets but few engines. Trends Plant Sci. 2013, 18, 505–513. [Google Scholar] [CrossRef]
- Heroes, E.; Lesage, B.; Görnemann, J.; Beullens, M.; Van Meervelt, L.; Bollen, M. The PP1 binding code: A molecular-lego strategy that governs specificity. FEBS J. 2013, 280, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Takemiya, A.; Kinoshita, T.; Asanuma, M.; Shimazaki, K.-I. Protein phosphatase 1 positively regulates stomatal opening in response to blue light in Vicia faba. Proc. Natl. Acad. Sci. USA 2006, 103, 13549–13554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradai, M.; Mahjoubi, H.; Chini, A.; Chabouté, M.E.; Hanin, M.; Ebel, C. Genome wide identification of wheat and Brachypodium type one protein phosphatases and functional characterization of durum wheat TdPP1a. PLoS ONE 2018, 13, e0191272. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, D.; Morita, H.; Hattori, T.; Takeda, S. Molecular characterization of the rice protein RSS1 required for meristematic activity under stressful conditions. Plant Physiol. Biochem. 2012, 61, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Wang, W.; Guo, X.; Yue, J.; Huang, Y.; Xu, X.; Li, J.; Hou, S. Arabidopsis DELLA Protein Degradation Is Controlled by a Type-One Protein Phosphatase, TOPP4. PLoS Genet. 2014, 10, e1004464. [Google Scholar] [CrossRef] [Green Version]
- Yue, J.; Qin, Q.; Meng, S.; Jing, H.; Gou, X.; Li, J.; Hou, S. TOPP4 regulates the stability of PHYTOCHROME INTERACTING FACTOR5 during photomorphogenesis in Arabidopsis. Plant Physiol. 2016, 170, 1381–1397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Qin, Q.; Yan, J.; Niu, Y.; Huang, B.; Guan, L.; Li, Y.; Ren, D.; Li, J.; Hou, S. TYPE-ONE PROTEIN PHOSPHATASE4 Regulates Pavement Cell Interdigitation by Modulating PIN-FORMED1 Polarity and Trafficking in Arabidopsis. Plant Physiol. 2015, 167, 1058–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Yan, J.; Qin, Q.; Zhang, J.; Chen, Y.; Zhao, L.; He, K.; Hou, S. Type one protein phosphatases (TOPPs) contribute to the plant defense response in Arabidopsis. J. Integr. Plant Biol. 2019, jipb.12845. [Google Scholar] [CrossRef]
- Yan, J.; Liu, Y.; Huang, X.; Li, L.; Hu, Z.; Zhang, J.; Qin, Q.; Yan, L.; He, K.; Wang, Y.; et al. An unreported NB-LRR protein SUT 1 is required for the autoimmune response mediated by type one protein phosphatase 4 mutation (topp4-1) in Arabidopsis. Plant J. 2019, 100, 357–373. [Google Scholar] [CrossRef]
- Templeton, G.W.; Nimick, M.; Morrice, N.; Campbell, D.; Goudreault, M.; Gingras, A.-C.; Takemiya, A.; Shimazaki, K.; Moorhead, G.B.G. Identification and characterization of AtI-2, an Arabidopsis homologue of an ancient protein phosphatase 1 (PP1) regulatory subunit. Biochem. J. 2011, 435, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.J.; Zhu, Y.; Wang, P.; Zhao, Y.; Xie, S.; Batelli, G.; Wang, B.; Duan, C.G.; Wang, X.; Xing, L.; et al. Type One Protein Phosphatase 1 and Its Regulatory Protein Inhibitor 2 Negatively Regulate ABA Signaling. PLoS Genet. 2016, 12, e1005835. [Google Scholar] [CrossRef] [Green Version]
- Franck, C.M.; Westermann, J.; Bürssner, S.; Lentz, R.; Lituiev, D.S.; Boisson-Dernier, A. The Protein Phosphatases ATUNIS1 and ATUNIS2 Regulate Cell Wall Integrity in Tip-Growing Cells. Plant Cell 2018, 30, 1906–1923. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, D.; Abe, K.; Miyao, A.; Kojima, M.; Sakakibara, H.; Mizutani, M.; Morita, H.; Toda, Y.; Hobo, T.; Sato, Y.; et al. RSS1 regulates the cell cycle and maintains meristematic activity under stress conditions in rice. Nat. Commun. 2011, 2, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.D.; Lin, K.H.; Chen, C.C.; Chiang, C.M. Oryza sativa protein phosphatase 1a (OsPP1a) involved in salt stress tolerance in transgenic rice. Mol. Breed. 2016, 36, 22. [Google Scholar] [CrossRef]
- Bollen, M.; Peti, W.; Ragusa, M.J.; Beullens, M. The extended PP1 toolkit: Designed to create specificity. Trends Biochem. Sci. 2010, 35, 450–458. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.K.H.; McCouch, S. Getting to the roots of it: Genetic and hormonal control of root architecture. Front. Plant Sci. 2013, 4, 186. [Google Scholar] [CrossRef] [Green Version]
- Ackerman-Lavert, M.; Savaldi-Goldstein, S. Growth models from a brassinosteroid perspective. Curr. Opin. Plant Biol. 2020, 53, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Garcia, M.-P.; Vilarrasa-Blasi, J.; Zhiponova, M.; Divol, F.; Mora-Garcia, S.; Russinova, E.; Cano-Delgado, A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 2011, 138, 849–859. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Li, J. Brassinosteroids Regulate Root Growth, Development, and Symbiosis. Mol. Plant 2016, 9, 86–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salazar-Henao, J.E.; Lehner, R.; Betegón-Putze, I.; Vilarrasa-Blasi, J.; Caño-Delgado, A.I. BES1 regulates the localization of the brassinosteroid receptor BRL3 within the provascular tissue of the Arabidopsis primary root. J. Exp. Bot. 2016, 67, 4951–4961. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Lu, D.; Gao, X.; Jiang, S.; Ma, X.; Wang, Z.; Mengiste, T.; Shan, L. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 2013, 110. [Google Scholar] [CrossRef] [Green Version]
- Leung, J.; Bouvier-Durand, M.; Morris, P.C.; Guerrier, D.; Chefdor, F.; Giraudat, J. Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 1994, 264, 1448–1452. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Leube, M.P.; Grill, E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 1994, 264, 1452–1455. [Google Scholar] [CrossRef]
- Pacifici, E.; Polverari, L.; Sabatini, S. Plant hormone cross-talk: The pivot of root growth. J. Exp. Bot. 2015, 66, 1113–1121. [Google Scholar] [CrossRef] [Green Version]
- Wachsman, G.; Sparks, E.E.; Benfey, P.N. Genes and networks regulating root anatomy and architecture. New Phytol. 2015, 208, 26–38. [Google Scholar] [CrossRef]
- Vukašinović, N.; Russinova, E. BRexit: Possible Brassinosteroid Export and Transport Routes. Trends Plant Sci. 2018, 23, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Vukašinović, N.; Wang, Y.; Vanhoutte, I.; Fendrych, M.; Guo, B.; Kvasnica, M.; Jiroutová, P.; Oklestkova, J.; Strnad, M.; Russinova, E. Local brassinosteroid biosynthesis enables optimal root growth. Nat. Plants 2021, 7, 619–632. [Google Scholar] [CrossRef]
- Chaiwanon, J.; Wang, Z.Y. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 2015, 25, 1031–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, L.; Zhang, A.; Wang, R.; Zhao, P.; Zhang, D.; Jiang, Y.; Diddugodage, C.J.; Wang, X.; Ni, Z.; Xu, S. Brassinosteroid Regulates Root Development with Highly Redundant Genes in Hexaploid Wheat. Plant Cell Physiol. 2019, 60, 1761–1777. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Li, L.; Zola, J.; Aluru, M.; Ye, H.; Foudree, A.; Guo, H.; Anderson, S.; Aluru, S.; Liu, P.; et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011, 65, 634–646. [Google Scholar] [CrossRef]
- Amorim-Silva, V.; García-Moreno, Á.; Castillo, A.G.; Lakhssassi, N.; Del Valle, A.E.; Pérez-Sancho, J.; Li, Y.; Posé, D.; Pérez-Rodriguez, J.; Lin, J.; et al. TTL proteins scaffold brassinosteroid signaling components at the plasma membrane to optimize signal transduction in arabidopsis. Plant Cell 2019, 31, 1807–1828. [Google Scholar] [CrossRef] [Green Version]
- Bojar, D.; Martinez, J.; Santiago, J.; Rybin, V.; Bayliss, R.; Hothorn, M. Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 2014, 78, 31–43. [Google Scholar] [CrossRef]
- Gehl, C.; Waadt, R.; Kudla, J.; Mendel, R.R.; Hänsch, R. New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant 2009, 2, 1051–1058. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ursache, R.; Andersen, T.G.; Marhavý, P.; Geldner, N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 2018, 93, 399–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bradai, M.; Amorim-Silva, V.; Belgaroui, N.; Esteban del Valle, A.; Chabouté, M.-E.; Schmit, A.-C.; Lozano-Duran, R.; Botella, M.A.; Hanin, M.; Ebel, C. Wheat Type One Protein Phosphatase Participates in the Brassinosteroid Control of Root Growth via Activation of BES1. Int. J. Mol. Sci. 2021, 22, 10424. https://doi.org/10.3390/ijms221910424
Bradai M, Amorim-Silva V, Belgaroui N, Esteban del Valle A, Chabouté M-E, Schmit A-C, Lozano-Duran R, Botella MA, Hanin M, Ebel C. Wheat Type One Protein Phosphatase Participates in the Brassinosteroid Control of Root Growth via Activation of BES1. International Journal of Molecular Sciences. 2021; 22(19):10424. https://doi.org/10.3390/ijms221910424
Chicago/Turabian StyleBradai, Mariem, Vitor Amorim-Silva, Nibras Belgaroui, Alicia Esteban del Valle, Marie-Edith Chabouté, Anne-Catherine Schmit, Rosa Lozano-Duran, Miguel Angel Botella, Moez Hanin, and Chantal Ebel. 2021. "Wheat Type One Protein Phosphatase Participates in the Brassinosteroid Control of Root Growth via Activation of BES1" International Journal of Molecular Sciences 22, no. 19: 10424. https://doi.org/10.3390/ijms221910424
APA StyleBradai, M., Amorim-Silva, V., Belgaroui, N., Esteban del Valle, A., Chabouté, M. -E., Schmit, A. -C., Lozano-Duran, R., Botella, M. A., Hanin, M., & Ebel, C. (2021). Wheat Type One Protein Phosphatase Participates in the Brassinosteroid Control of Root Growth via Activation of BES1. International Journal of Molecular Sciences, 22(19), 10424. https://doi.org/10.3390/ijms221910424