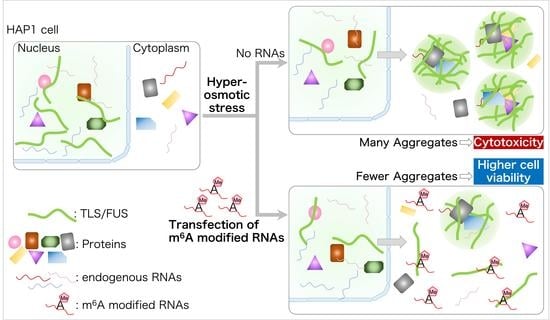
m6A Modified Short RNA Fragments Inhibit Cytoplasmic TLS/FUS Aggregation Induced by Hyperosmotic Stress
Abstract
:1. Introduction
2. Results
2.1. TLS/FUS Binds Intensely to m6A-Modified RNA Fragments
2.2. m6A-Modified RNA Fragments Did Not Promote LLPS of TLS/FUS
2.3. Cytoplasmic TLS/FUS Aggregation Is Inhibited by m6A-Modified RNA Transfection
2.4. m6A-Modified RNA Fragment Has a Different Effect on Localization of TLS/FUS-Interacting Proteins
2.5. m6A-Modified RNA Fragments Enhance the Viability of Cells Treated with Sorbitol
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Stable Cell Preparation
4.2. RNA Pull Down Assay and Overexpression of GFP-TLS/FUS
4.3. Western Blot Analysis
4.4. Immunocytochemistry (ICC) Assay
4.5. RNA Induction and Sorbitol Treatment
4.6. Cell Viability Assay
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shang, Y.; Huang, E.J. Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis. Brain Res. 2016, 1647, 65–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L. FUS mutation is probably the most common pathogenic gene for JALS, especially sporadic JALS. Rev. Neurol. 2021, 177, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Aksoy, Y.A.; Deng, W.; Stoddart, J.; Chung, R.; Guillemin, G.; Cole, N.J.; Neely, G.G.; Hesselson, D. “STRESSED OUT”: The role of FUS and TDP-43 in amyotrophic lateral sclerosis. Int. J. Biochem. Cell Biol. 2020, 126, 105821. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Lyashchenko, A.K.; Lu, L.; Nasrabady, S.E.; Elmaleh, M.; Mendelsohn, M.; Nemes, A.; Tapia, J.C.; Mentis, G.Z.; Shneider, N.A. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat. Commun. 2016, 7, 10465. [Google Scholar] [CrossRef] [PubMed]
- Pakravan, D.; Michiels, E.; Bratek-Skicki, A.; De Decker, M.; Van Lindt, J.; Alsteens, D.; Derclaye, S.; Van Damme, P.; Schymkowitz, J.; Rousseau, F.; et al. Liquid–Liquid Phase Separation Enhances TDP-43 LCD Aggregation but Delays Seeded Aggregation. Biomolecules 2021, 11, 548. [Google Scholar] [CrossRef]
- Tsoi, P.S.; Quan, M.D.; Choi, K.-J.; Dao, K.M.; Ferreon, J.C.; Ferreon, A.C.M. Electrostatic modulation of hnRNPA1 low-complexity domain liquid–liquid phase separation and aggregation. Protein Sci. 2021, 30, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.S.; Harrell, L.M.; Wieland, D.R.; Lay, M.A.; Thompson, V.F.; Schwartz, J.C. Fusion protein EWS-FLI1 is incorporated into a protein granule in cells. RNA 2021, 27, 920–932. [Google Scholar] [CrossRef]
- Berkeley, R.F.; Kashefi, M.; Debelouchina, G.T. Real-time observation of structure and dynamics during the liquid-to-solid transition of FUS LC. Biophys. J. 2021, 120, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Gomes, E.; Shorter, J. The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294, 7115–7127. [Google Scholar] [CrossRef] [Green Version]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef] [Green Version]
- Costa, I.D.; Buchanan, C.N.; Zdradzinski, M.D.; Sahoo, P.K.; Smith, T.P.; Thames, E.; Kar, A.N.; Twiss, J.L. The functional organization of axonal mRNA transport and translation. Nat. Rev. Neurosci. 2021, 22, 77–91. [Google Scholar] [CrossRef]
- Darling, A.L.; Shorter, J. Combating deleterious phase transitions in neurodegenerative disease. Biochim. Biophys. Acta BBA Mol. Cell Res. 2021, 1868, 118984. [Google Scholar] [CrossRef] [PubMed]
- George-Hyslop, P.S.; Lin, J.Q.; Miyashita, A.; Phillips, E.C.; Qamar, S.; Randle, S.J.; Wang, G. The physiological and pathological biophysics of phase separation and gelation of RNA binding proteins in amyotrophic lateral sclerosis and fronto-temporal lobar degeneration. Brain Res. 2018, 1693, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Bogaert, E.; Boeynaems, S.; Kato, M.; Guo, L.; Caulfield, T.R.; Steyaert, J.; Scheveneels, W.; Wilmans, N.; Haeck, W.; Hersmus, N.; et al. Molecular Dissection of FUS Points at Synergistic Effect of Low-Complexity Domains in Toxicity. Cell Rep. 2018, 24, 529–537.e4. [Google Scholar] [CrossRef] [Green Version]
- Najafi, S.; Lin, Y.; Longhini, A.P.; Zhang, X.; Delaney, K.T.; Kosik, K.S.; Fredrickson, G.H.; Shea, J.-E.; Han, S. Liquid–liquid phase separation of Tau by self and complex coacervation. Protein Sci. 2021, 30, 1393–1407. [Google Scholar] [CrossRef] [PubMed]
- Boyko, S.; Surewicz, K.; Surewicz, W.K. Regulatory mechanisms of tau protein fibrillation under the conditions of liquid–liquid phase separation. Proc. Natl. Acad. Sci. USA 2020, 117, 31882–31890. [Google Scholar] [CrossRef] [PubMed]
- Birsa, N.; Bentham, M.P.; Fratta, P. Cytoplasmic functions of TDP-43 and FUS and their role in ALS. Semin. Cell Dev. Biol. 2020, 99, 193–201. [Google Scholar] [CrossRef]
- Lattante, S.; Rouleau, G.A.; Kabashi, E. TARDBP and FUS Mutations Associated with Amyotrophic Lateral Sclerosis: Summary and Update. Hum. Mutat. 2013, 34, 812–826. [Google Scholar] [CrossRef] [PubMed]
- Qamar, S.; Wang, G.; Randle, S.J.; Ruggeri, F.S.; Varela, J.A.; Lin, J.Q.; Phillips, E.C.; Miyashita, A.; Williams, D.; Ströhl, F.; et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions. Cell 2018, 173, 720–734.e15. [Google Scholar] [CrossRef] [Green Version]
- Monahan, Z.; Ryan, V.H.; Janke, A.M.; Burke, K.A.; Rhoads, S.N.; Zerze, G.H.; O’Meally, R.; Dignon, G.L.; Conicella, A.E.; Zheng, W.; et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017, 36, 2951–2967. [Google Scholar] [CrossRef]
- Hamad, N.; Yoneda, R.; So, M.; Kurokawa, R.; Nagata, T.; Katahira, M. Non-coding RNA suppresses FUS aggregation caused by mechanistic shear stress on pipetting in a sequence-dependent manner. Sci. Rep. 2021, 11, 9523. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, T.; Ali, R.; Jiou, J.; Fung, H.Y.J.; Burke, K.A.; Kim, S.J.; Lin, Y.; Peeples, W.B.; Saltzberg, D.; Soniat, M.; et al. Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell 2018, 173, 693–705.e22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Zhang, S.; Gu, J.; Tong, Y.; Li, Y.; Gui, X.; Long, H.; Wang, C.; Zhao, C.; Lu, J.; et al. Hsp27 chaperones FUS phase separation under the modulation of stress-induced phosphorylation. Nat. Struct. Mol. Biol. 2020, 27, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Maharana, S.; Wang, J.; Papadopoulos, D.K.; Richter, D.; Pozniakovsky, A.; Poser, I.; Bickle, M.; Rizk, S.; Guillén-Boixet, J.; Franzmann, T.M.; et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 2018, 360, 918–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levone, B.R.; Lenzken, S.C.; Antonaci, M.; Maiser, A.; Rapp, A.; Conte, F.; Reber, S.; Mechtersheimer, J.; Ronchi, A.E.; Mühlemann, O.; et al. FUS-dependent liquid–liquid phase separation is important for DNA repair initiation. J. Cell Biol. 2021, 220. [Google Scholar] [CrossRef]
- Widagdo, J.; Anggono, V. The m6A-epitranscriptomic signature in neurobiology: From neurodevelopment to brain plasticity. J. Neurochem. 2018, 147, 137–152. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, L.; Diao, J.; Shi, Y.G.; Shi, Y.; Ma, H.; Shen, H. Binding to m6A RNA promotes YTHDF2-mediated phase separation. Protein Cell 2020, 11, 304–307. [Google Scholar] [CrossRef] [Green Version]
- Hamad, N.; Mashima, T.; Yamaoki, Y.; Kondo, K.; Yoneda, R.; Oyoshi, T.; Kurokawa, R.; Nagata, T.; Katahira, M. RNA sequence and length contribute to RNA-induced conformational change of TLS/FUS. Sci. Rep. 2020, 10, 2629. [Google Scholar] [CrossRef]
- Yoneda, R.; Suzuki, S.; Mashima, T.; Kondo, K.; Nagata, T.; Katahira, M.; Kurokawa, R. The binding specificity of Translocated in LipoSarcoma/FUsed in Sarcoma with lncRNA transcribed from the promoter region of cyclin D1. Cell Biosci. 2016, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Yoneda, R.; Ueda, N.; Uranishi, K.; Hirasaki, M.; Kurokawa, R. Long noncoding RNA pncRNA-D reduces cyclin D1 gene expression and arrests cell cycle through RNA m6A modification. J. Biol. Chem. 2020, 295, 5626–5639. [Google Scholar] [CrossRef] [Green Version]
- Ajmone-Cat, M.A.; Onori, A.; Toselli, C.; Stronati, E.; Morlando, M.; Bozzoni, I.; Monni, E.; Kokaia, Z.; Lupo, G.; Minghetti, L.; et al. Increased FUS levels in astrocytes leads to astrocyte and microglia activation and neuronal death. Sci. Rep. 2019, 9, 4572. [Google Scholar] [CrossRef] [PubMed]
- Sama, R.R.K.; Ward, C.L.; Kaushansky, L.J.; Lemay, N.; Ishigaki, S.; Urano, F.; Bosco, D.A. FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. J. Cell. Physiol. 2013, 228, 2222–2231. [Google Scholar] [CrossRef] [Green Version]
- Baron, D.M.; Kaushansky, L.J.; Ward, C.L.; Sama, R.R.K.; Chian, R.-J.; Boggio, K.J.; Quaresma, A.J.C.; Nickerson, J.A.; Bosco, D.A. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol. Neurodegener. 2013, 8, 30. [Google Scholar] [CrossRef] [Green Version]
- Takanashi, K.; Yamaguchi, A. Aggregation of ALS-linked FUS mutant sequesters RNA binding proteins and impairs RNA granules formation. Biochem. Biophys. Res. Commun. 2014, 452, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Farrawell, N.E.; Lambert-Smith, I.A.; Warraich, S.T.; Blair, I.P.; Saunders, D.N.; Hatters, D.M.; Yerbury, J.J. Distinct partitioning of ALS associated TDP-43, FUS and SOD1 mutants into cellular inclusions. Sci. Rep. 2015, 5, 13416. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Takanashi, K. FUS interacts with nuclear matrix-associated protein SAFB1 as well as Matrin3 to regulate splicing and ligand-mediated transcription. Sci. Rep. 2016, 6, 35195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dormann, D.; Rodde, R.; Edbauer, D.; Bentmann, E.; Fischer, I.; Hruscha, A.; Than, M.E.; Mackenzie, I.R.A.; Capell, A.; Schmid, B.; et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010, 29, 2841–2857. [Google Scholar] [CrossRef] [Green Version]
- Hoell, J.I.; Larsson, E.; Runge, S.; Nusbaum, J.D.; Duggimpudi, S.; Farazi, T.A.; Hafner, M.; Borkhardt, A.; Sander, C.; Tuschl, T. RNA targets of wild-type and mutant FET family proteins. Nat. Struct. Mol. Biol. 2011, 18, 1428–1431. [Google Scholar] [CrossRef]
- Reber, S.; Jutzi, D.; Lindsay, H.; Devoy, A.; Mechtersheimer, J.; Levone, B.R.; Domanski, M.; Bentmann, E.; Dormann, D.; Mühlemann, O.; et al. The phase separation-dependent FUS interactome reveals nuclear and cytoplasmic function of liquid–liquid phase separation. Nucleic Acids Res. 2021, 49, 7713–7731. [Google Scholar] [CrossRef]
- Zhu, T.; Roundtree, I.A.; Wang, P.; Wang, X.; Wang, L.; Sun, C.; Tian, Y.; Li, J.; He, C.; Xu, Y. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014, 24, 1493–1496. [Google Scholar] [CrossRef] [Green Version]
- Shi, R.; Ying, S.; Li, Y.; Zhu, L.; Wang, X.; Jin, H. Linking the YTH domain to cancer: The importance of YTH family proteins in epigenetics. Cell Death Dis. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Hazra, D.; Chapat, C.; Graille, M. m6A mRNA Destiny: Chained to the rhYTHm by the YTH-Containing Proteins. Genes 2019, 10, 49. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Pei, G.; Li, D.; Li, R.; Shao, Y.; Zhang, Q.C.; Li, P. Multivalent m6A motifs promote phase separation of YTHDF proteins. Cell Res. 2019, 29, 767–769. [Google Scholar] [CrossRef]
- Liu, S.-Y.; Feng, Y.; Wu, J.-J.; Zou, M.-L.; Sun, Z.-L.; Li, X.; Yuan, F.-L. m 6 A facilitates YTHDF-independent phase separation. J. Cell. Mol. Med. 2020, 24, 2070–2072. [Google Scholar] [CrossRef] [Green Version]
- Hamad, N.; Watanabe, H.; Uchihashi, T.; Kurokawa, R.; Nagata, T.; Katahira, M. Direct visualization of the conformational change of FUS/TLS upon binding to promoter-associated non-coding RNA. Chem. Commun. 2020, 56, 9134–9137. [Google Scholar] [CrossRef]
- Johnson, J.O.; Pioro, E.P.; Boehringer, A.; Chia, R.; Feit, H.; Renton, A.E.; Pliner, H.A.; Abramzon, Y.; Marangi, G.; Winborn, B.J.; et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 2014, 17, 664–666. [Google Scholar] [CrossRef]
- Lin, K.-P.; Tsai, P.-C.; Liao, Y.-C.; Chen, W.-T.; Tsai, C.-P.; Soong, B.-W.; Lee, Y.-C. Mutational analysis of MATR3 in Taiwanese patients with amyotrophic lateral sclerosis. Neurobiol. Aging 2015, 36, 2005.e1–2005.e4. [Google Scholar] [CrossRef] [PubMed]
- Tada, M.; Doi, H.; Koyano, S.; Kubota, S.; Fukai, R.; Hashiguchi, S.; Hayashi, N.; Kawamoto, Y.; Kunii, M.; Tanaka, K.; et al. Matrin 3 Is a Component of Neuronal Cytoplasmic Inclusions of Motor Neurons in Sporadic Amyotrophic Lateral Sclerosis. Am. J. Pathol. 2018, 188, 507–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogelj, B.; Easton, L.E.; Bogu, G.K.; Stanton, L.W.; Rot, G.; Curk, T.; Zupan, B.; Sugimoto, Y.; Modic, M.; Haberman, N.; et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci. Rep. 2012, 2, 603. [Google Scholar] [CrossRef] [PubMed]
- Sreedharan, J.; Blair, I.P.; Tripathi, V.B.; Hu, X.; Vance, C.; Rogelj, B.; Ackerley, S.; Durnall, J.C.; Williams, K.L.; Buratti, E.; et al. TDP-43 Mutations in Familial and Sporadic Amyotrophic Lateral Sclerosis. Science 2008, 319, 1668–1672. [Google Scholar] [CrossRef] [PubMed]
- Ulke-Lemée, A.; Trinkle-Mulcahy, L.; Chaulk, S.; Bernstein, N.K.; Morrice, N.; Glover, M.; Lamond, A.I.; Moorhead, G.B.G. The nuclear PP1 interacting protein ZAP3 (ZAP) is a putative nucleoside kinase that complexes with SAM68, CIA, NF110/45, and HNRNP-G. Biochim. Biophys. Acta BBA Proteins Proteom. 2007, 1774, 1339–1350. [Google Scholar] [CrossRef]
- Schwartz, J.C.; Podell, E.R.; Han, S.S.W.; Berry, J.D.; Eggan, K.C.; Cech, T.R. FUS is sequestered in nuclear aggregates in ALS patient fibroblasts. Mol. Biol. Cell 2014, 25, 2571–2578. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, N.; Pandey, U.B. Autophagy Dysregulation in ALS: When Protein Aggregates Get Out of Hand. Front. Mol. Neurosci. 2017, 10, 263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroschwald, S.; Maharana, S.; Simon, A. Hexanediol: A chemical probe to investigate the material properties of membrane-less compartments. Matters 2017, 3. [Google Scholar] [CrossRef] [Green Version]
- Ueda, N.; Hirose, Y.; Yoneda, R.; Bando, T.; Kurokawa, R. Potential Inhibitor Against Phase Separation, 1,6-hexanediol Specifically Binds to Beta Actin in Nuclear Extract of Human Cell Line. Biomed. Sci. 2020, 6, 89. [Google Scholar] [CrossRef]
- Düster, R.; Kaltheuner, I.H.; Schmitz, M.; Geyer, M. 1,6-Hexanediol, commonly used to dissolve liquid–liquid phase separated condensates, directly impairs kinase and phosphatase activities. J. Biol. Chem. 2021, 296, 100260. [Google Scholar] [CrossRef]
- Itoh, Y.; Iida, S.; Tamura, S.; Nagashima, R.; Shiraki, K.; Goto, T.; Hibino, K.; Ide, S.; Maeshima, K. 1,6-hexanediol rapidly immobilizes and condenses chromatin in living human cells. Life Sci. Alliance 2021, 4, e202001005. [Google Scholar] [CrossRef]
- Khorkova, O.; Hsiao, J.; Wahlestedt, C. Basic biology and therapeutic implications of lncRNA. Adv. Drug Deliv. Rev. 2015, 87, 15–24. [Google Scholar] [CrossRef] [Green Version]
- Blokhin, I.; Khorkova, O.; Hsiao, J.; Wahlestedt, C. Developments in lncRNA drug discovery: Where are we heading? Expert Opin. Drug Discov. 2018, 13, 837–849. [Google Scholar] [CrossRef]
- Zhou, C.-C.; Yang, F.; Yuan, S.-X.; Ma, J.-Z.; Liu, F.; Yuan, J.-H.; Bi, F.-R.; Lin, K.-Y.; Yin, J.-H.; Cao, G.-W.; et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology 2016, 63, 850–863. [Google Scholar] [CrossRef]
- Scoles, D.R.; Pulst, S.M. Oligonucleotide therapeutics in neurodegenerative diseases. RNA Biol. 2018, 15, 707–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, W.; Yoneda, R.; Ueda, N.; Kurokawa, R. Arginine methylation of translocated in liposarcoma (TLS) inhibits its binding to long noncoding RNA, abrogating TLS-mediated repression of CBP/p300 activity. J. Biol. Chem. 2018, 293, 10937–10948. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoneda, R.; Ueda, N.; Kurokawa, R. m6A Modified Short RNA Fragments Inhibit Cytoplasmic TLS/FUS Aggregation Induced by Hyperosmotic Stress. Int. J. Mol. Sci. 2021, 22, 11014. https://doi.org/10.3390/ijms222011014
Yoneda R, Ueda N, Kurokawa R. m6A Modified Short RNA Fragments Inhibit Cytoplasmic TLS/FUS Aggregation Induced by Hyperosmotic Stress. International Journal of Molecular Sciences. 2021; 22(20):11014. https://doi.org/10.3390/ijms222011014
Chicago/Turabian StyleYoneda, Ryoma, Naomi Ueda, and Riki Kurokawa. 2021. "m6A Modified Short RNA Fragments Inhibit Cytoplasmic TLS/FUS Aggregation Induced by Hyperosmotic Stress" International Journal of Molecular Sciences 22, no. 20: 11014. https://doi.org/10.3390/ijms222011014
APA StyleYoneda, R., Ueda, N., & Kurokawa, R. (2021). m6A Modified Short RNA Fragments Inhibit Cytoplasmic TLS/FUS Aggregation Induced by Hyperosmotic Stress. International Journal of Molecular Sciences, 22(20), 11014. https://doi.org/10.3390/ijms222011014