Aggregation and Its Influence on the Bioactivities of a Novel Antimicrobial Peptide, Temporin-PF, and Its Analogues
Abstract
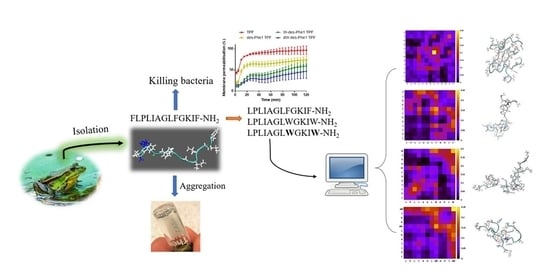
:1. Introduction
2. Results
2.1. Identification and Characterization of Temporin-PF from the Skin Secretion
2.2. Peptide Synthesis and Characterization
2.3. Observation of Aggregation Effect of Synthetic Peptides
2.4. MD Simulation of Aggregation Effect of Peptides
2.5. Secondary Structure Analysis of TPF and the Analogs
2.6. Antimicrobial, Antibiofilm, and Hemolytic Activity of TPE and the Analogs
2.7. S. aureus Membrane Permeabilization
3. Discussion
4. Materials and Methods
4.1. Specimen Biodata and Harvesting of Skin Secretion
4.2. Identification of Precursor-Encoding cDNA from the Skin Secretion
4.3. Chemical Peptide Synthesis
4.4. Observation of Aggregation of The Peptides
4.5. Molecular Dynamic Simulation
4.6. Circular Dichorism
4.7. Antimicrobial Assays
4.8. Determination of Minimal Biofilm Inhibitory Concentration (MBIC) and Minimal Biofilm Eradication Concentration (MBEC)
4.9. Hemolysis Test
4.10. SYTOX Green Stain Uptake Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mangoni, M.L.; Di Grazia, A.; Cappiello, F.; Casciaro, B.; Luca, V. Naturally Occurring Peptides from Rana temporaria: Antimicrobial Properties and More. Curr. Top. Med. Chem. 2016, 16, 54–64. [Google Scholar] [CrossRef]
- Lyttle, T.; Goldstein, D.; Gartz, J. Bufo toads and bufotenine: Fact and fiction surrounding an alleged psychedelic. J. Psychoact. Drugs 1996, 28, 267–290. [Google Scholar] [CrossRef]
- Daly, J.W.; Garraffo, H.M.; Spande, T.F.; Decker, M.W.; Sullivan, J.P.; Williams, M. Alkaloids from frog skin: The discovery of epibatidine and the potential for developing novel non-opioid analgesics. Nat. Prod. Rep. 2000, 17, 131–135. [Google Scholar] [CrossRef] [Green Version]
- Conlon, J.M. Structural diversity and species distribution of host-defense peptides in frog skin secretions. Cell. Mol. Life Sci. 2011, 68, 2303–2315. [Google Scholar] [CrossRef]
- Diamond, G.; Beckloff, N.; Weinberg, A.; Kisich, K.O. The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. 2009, 15, 2377–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raaymakers, C.; Verbrugghe, E.; Hernot, S.; Hellebuyck, T.; Betti, C.; Peleman, C.; Claeys, M.; Bert, W.; Caveliers, V.; Ballet, S.; et al. Antimicrobial peptides in frog poisons constitute a molecular toxin delivery system against predators. Nat. Commun. 2017, 8, 1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, X.; Dong, F.; Shi, C.; Liu, S.; Sun, J.; Chen, J.; Li, H.; Xu, H.; Lao, X.; Zheng, H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 2019, 6, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rai, D.K.; Qian, S. Interaction of the Antimicrobial Peptide Aurein 1.2 and Charged Lipid Bilayer. Sci. Rep. 2017, 7, 3719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Pan, H.; Wang, H.; Liao, X.; Sun, D.; Xu, K.; Chen, T.; Zhang, X.; Wu, M.; Wu, D.; et al. Identification of new dermaseptins with self-assembly tendency: Membrane disruption, biofilm eradication, and infected wound healing efficacy. Acta Biomater. 2020, 109, 208–219. [Google Scholar] [CrossRef]
- Proaño-Bolaños, C.; Blasco-Zúñiga, A.; Almeida, J.R.; Wang, L.; Llumiquinga, M.A.; Rivera, M.; Zhou, M.; Chen, T.; Shaw, C. Unravelling the Skin Secretion Peptides of the Gliding Leaf Frog, Agalychnis spurrelli (Hylidae). Biomolecules 2019, 9, 667. [Google Scholar] [CrossRef] [Green Version]
- Amiche, M.; Ladram, A.; Nicolas, P. A consistent nomenclature of antimicrobial peptides isolated from frogs of the subfamily Phyllomedusinae. Peptides 2008, 29, 2074–2082. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, D.; Wang, L.; Lin, C.; Ma, C.; Xi, X.; Zhou, M.; Duan, J.; Bininda-Emonds, O.R.P.; Chen, T. Targeted modification of a novel amphibian antimicrobial peptide from Phyllomedusa tarsius to enhance its activity against MRSA and microbial biofilm. Front. Microbiol. 2017, 8, 628. [Google Scholar] [CrossRef] [Green Version]
- Ghavami, S.; Asoodeh, A.; Klonisch, T.; Halayko, A.J.; Kadkhoda, K.; Kroczak, T.J.; Gibson, S.B.; Booy, E.P.; Naderi-Manesh, H.; Los, M. Brevinin-2R1 semi-selectively kills cancer cells by a distinct mechanism, which involves the lysosomal-mitochondrial death pathway. J. Cell. Mol. Med. 2008, 12, 1005–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponti, D.; Mignogna, G.; Mangoni, M.L.; De Biase, D.; Simmaco, M.; Barra, D. Expression and activity of cyclic and linear analogues of esculentin-1, an anti-microbial peptide from amphibian skin. Eur. J. Biochem. 1999, 263, 921–927. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Zai, Y.; Xi, X.; Ma, C.; Wang, L.; Zhou, M.; Shaw, C.; Chen, T. BBA—General Subjects A novel membrane-disruptive antimicrobial peptide from frog skin secretion against cystic fi brosis isolates and evaluation of anti-MRSA effect using Galleria mellonella model. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 849–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, K.; Yuan, W.; Ma, C.; Yu, X.; Wang, L.; Hong, M.; Xi, X.; Zhou, M.; Chen, T. Modification targeting the “Rana Box” motif of a novel nigrocin peptide from Hylarana latouchii enhances and broadens its potency against multiple bacteria. Front. Microbiol. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Kolodziejek, J.; Mechkarska, M.; Coquet, L.; Leprince, J.; Jouenne, T.; Vaudry, H.; Nielsen, P.F.; Nowotny, N.; King, J.D. Host defense peptides from Lithobates forreri, Hylarana luctuosa, and Hylarana signata (Ranidae): Phylogenetic relationships inferred from primary structures of ranatuerin-2 and brevinin-2 peptides. Comp. Biochem. Physiol. Part D Genom. Proteom. 2014, 9, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Shi, D.; Zhong, R.; Ye, Z.; Ma, C.; Zhou, M.; Xi, X.; Wang, L.; Chen, T.; Kwok, H.F. Bioevaluation of ranatuerin-2Pb from the frog skin secretion of Rana pipiens and its truncated analogues. Biomolecules 2019, 9, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savelyeva, A.; Ghavami, S.; Davoodpour, P.; Asoodeh, A.; Łos, M.J. An overview of brevinin superfamily: Structure, function and clinical perspectives. Adv. Exp. Med. Biol. 2014, 818, 197–212. [Google Scholar]
- Sang, M.; Wu, Q.; Xi, X.; Ma, C.; Wang, L.; Zhou, M.; Burrows, J.F.; Chen, T. Identification and target-modifications of temporin-PE: A novel antimicrobial peptide in the defensive skin secretions of the edible frog, Pelophylax kl. esculentus. Biochem. Biophys. Res. Commun. 2017, 495, 2539–2546. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; He, H.; Chen, X.; Zhou, M.; Wei, M.; Xi, X.; Ma, C.; Du, Q.; Chen, T.; Shaw, C.; et al. A Novel Antimicrobial Peptide (Kassinatuerin-3) Isolated from the Skin Secretion of the African Frog, Kassina senegalensis. Biology 2020, 9, 148. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 1987, 84, 5449–5453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangoni, M.L. Temporins, anti-infective peptides with expanding properties. Cell. Mol. Life Sci. 2006, 63, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Simmaco, M.; Mignogna, G.; Canofeni, S.; Miele, R.; Mangoni, M.L.; Barra, D. Temporins, antimicrobial peptides from the European red frog Rana temporaria. Eur. J. Biochem. 1996, 242, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Mangoni, M.L.; Shai, Y. Short native antimicrobial peptides and engineered ultrashort lipopeptides: Similarities and differences in cell specificities and modes of action. Cell. Mol. Life Sci. 2011, 68, 2267–2280. [Google Scholar] [CrossRef]
- Xie, Z.; Wei, H.; Meng, J.; Cheng, T.; Song, Y.; Wang, M.; Zhang, Y. The Analogs of Temporin-GHa Exhibit a Broader Spectrum of Antimicrobial Activity and a Stronger Antibiofilm Potential against Staphylococcus aureus. Molecules 2019, 24, 4173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makovitzki, A.; Avrahami, D.; Shai, Y. Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. USA 2006, 103, 15997–16002. [Google Scholar] [CrossRef] [Green Version]
- Mishra, B.; Wang, X.; Lushnikova, T.; Zhang, Y.; Golla, R.M.; Narayana, J.L.; Wang, C.; McGuire, T.R.; Wang, G. Antibacterial, antifungal, anticancer activities and structural bioinformatics analysis of six naturally occurring temporins. Peptides 2018, 106, 9–20. [Google Scholar] [CrossRef]
- Crépin, A.; Jégou, J.-F.; André, S.; Ecale, F.; Croitoru, A.; Cantereau, A.; Berjeaud, J.-M.; Ladram, A.; Verdon, J. In vitro and intracellular activities of frog skin temporins against Legionella pneumophila and its eukaryotic hosts. Sci. Rep. 2020, 10, 3978. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, S.; Ghosh, J.K. Introduction of a lysine residue promotes aggregation of temporin L in lipopolysaccharides and augmentation of its antiendotoxin property. Antimicrob. Agents Chemother. 2013, 57, 2457–2466. [Google Scholar] [CrossRef] [Green Version]
- Marcocci, M.E.; Amatore, D.; Villa, S.; Casciaro, B.; Aimola, P.; Franci, G.; Grieco, P.; Galdiero, M.; Palamara, A.T.; Mangoni, M.L.; et al. The Amphibian Antimicrobial Peptide Temporin B Inhibits In Vitro Herpes Simplex Virus 1 Infection. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Mangoni, M.L.; Carotenuto, A.; Auriemma, L.; Saviello, M.R.; Campiglia, P.; Gomez-Monterrey, I.; Malfi, S.; Marcellini, L.; Barra, D.; Novellino, E.; et al. Structure-activity relationship, conformational and biological studies of temporin L analogues. J. Med. Chem. 2011, 54, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Mohanram, H.; Bhattacharjya, S. Resurrecting inactive antimicrobial peptides from the lipopolysaccharide trap. Antimicrob. Agents Chemother. 2014, 58, 1987–1996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, H.; Xie, Z.; Wei, H.; Zhang, S.; Song, Y.; Wang, M.; Zhang, Y. Antibacterial and Antibiofilm Activity of Temporin-GHc and Temporin-GHd Against Cariogenic Bacteria, Streptococcus mutans. Front. Microbiol. 2019, 10, 2854. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, S.; Kumar, A.; Tripathi, A.K.; Tandon, A.; Ghosh, J.K. Modulation of anti-endotoxin property of Temporin L by minor amino acid substitution in identified phenylalanine zipper sequence. Biochem. J. 2016, 473, 4045–4062. [Google Scholar] [CrossRef]
- Saravanan, R.; Joshi, M.; Mohanram, H.; Bhunia, A.; Mangoni, M.L.; Bhattacharjya, S. NMR Structure of Temporin-1 Ta in Lipopolysaccharide Micelles: Mechanistic Insight into Inactivation by Outer Membrane. PLoS ONE 2013, 8, e72718. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.U.; Rehman, A.U.; Liu, H.; Chen, H.F. Comparison and evaluation of force fields for intrinsically disordered proteins. J. Chem. Inf. Model. 2020, 60, 4912–4923. [Google Scholar] [CrossRef] [PubMed]
- Piana, S.; Donchev, A.G.; Robustelli, P.; Shaw, D.E. Water dispersion interactions strongly influence simulated structural properties of disordered protein states. J. Phys. Chem. B 2015, 119, 5113–5123. [Google Scholar] [CrossRef]
- Farrotti, A.; Conflitti, P.; Srivastava, S.; Ghosh, J.K.; Palleschi, A.; Stella, L.; Bocchinfuso, G. Molecular Dynamics Simulations of the Host Defense Peptide Temporin L and Its Q3K Derivative: An Atomic Level View from Aggregation in Water to Bilayer Perturbation. Molecules 2017, 22, 1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzo, G.; Ferguson, P.M.; Hind, C.K.; Clifford, M.; Gustilo, V.B.; Ali, H.; Bansal, S.S.; Bui, T.T.; Drake, A.F.; Atkinson, R.A.; et al. Temporin L and aurein 2.5 have identical conformations but subtly distinct membrane and antibacterial activities. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Bellavita, R.; Falanga, A.; Buommino, E.; Merlino, F.; Casciaro, B.; Cappiello, F.; Mangoni, M.L.; Novellino, E.; Catania, M.R.; Paolillo, R.; et al. Novel temporin L antimicrobial peptides: Promoting self-assembling by lipidic tags to tackle superbugs. J. Enzym. Inhib. Med. Chem. 2020, 35, 1751–1764. [Google Scholar] [CrossRef] [PubMed]
- Sarig, H.; Rotem, S.; Ziserman, L.; Danino, D.; Mor, A. Impact of self-assembly properties on antibacterial activity of short acyl-lysine oligomers. Antimicrob. Agents Chemother. 2008, 52, 4308–4314. [Google Scholar] [CrossRef] [Green Version]
- Haney, E.F.; Wu, B.C.; Lee, K.; Hilchie, A.L.; Hancock, R.E.W. Aggregation and Its Influence on the Immunomodulatory Activity of Synthetic Innate Defense Regulator Peptides. Cell Chem. Biol. 2017, 24, 969–980. [Google Scholar] [CrossRef]
- Oren, Z.; Lerman, J.C.; Gudmundsson, G.H.; Agerberth, B.; Shai, Y. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: Relevance to the molecular basis for its non-cell-selective activity. Biochem. J. 1999, 341, 501–513. [Google Scholar] [CrossRef]
- Feder, R.; Dagan, A.; Mor, A. Structure-activity relationship study of antimicrobial dermaseptin S4 showing the consequences of peptide oligomerization on selective cytotoxicity. J. Biol. Chem. 2000, 275, 4230–4238. [Google Scholar] [CrossRef] [Green Version]
- Skibiszewska, S.; Żaczek, S.; Dybala-Defratyka, A.; Jędrzejewska, K.; Jankowska, E. Influence of short peptides with aromatic amino acid residues on aggregation properties of serum amyloid A and its fragments. Arch. Biochem. Biophys. 2020, 681. [Google Scholar] [CrossRef] [PubMed]
- Dennison, S.R.; Whittaker, M.; Harris, F.; Phoenix, D.A. Anticancer alpha-helical peptides and structure/function relationships underpinning their interactions with tumour cell membranes. Curr. Protein Pept. Sci. 2006, 7, 487–499. [Google Scholar] [CrossRef]
- Du, D.; Bunagan, M.R.; Gai, F. The effect of charge-charge interactions on the kinetics of α-helix formation. Biophys. J. 2007, 93, 4076–4082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottler, L.M.; Ramamoorthy, A. Structure, membrane orientation, mechanism, and function of pexiganan—A highly potent antimicrobial peptide designed from magainin. Biochim. Biophys. Acta Biomembr. 2009, 1788, 1680–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.H.; Melo, M.C.; Berglund, N.; Khan, A.; de la Fuente, C.; Ulmschneider, J.P.; Ulmschneider, M.B. Understanding and modelling the interactions of peptides with membranes: From partitioning to self-assembly. Curr. Opin. Struct. Biol. 2020, 61, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Peptides, A. Tryptophan-Rich and Proline-Rich Antimicrobial Peptides. Molecules 2018, 23, 815. [Google Scholar] [CrossRef] [Green Version]
- Rozek, A.; Powers, J.P.S.; Friedrich, C.L.; Hancock, R.E.W. Structure-Based Design of an Indolicidin Peptide Analogue with Increased Protease Stability. Biochemistry 2003, 42, 14130–14138. [Google Scholar] [CrossRef] [PubMed]
- Lequin, O.; Bruston, F.; Convert, O.; Chassaing, G.; Nicolas, P. Helical structure of dermaseptin B2 in a membrane-mimetic environment. Biochemistry 2003, 42, 10311–10323. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, D.; Xi, X.; Wu, Y.; Ma, C.; Zhou, M.; Wang, L.; Yang, M.; Chen, T.; Shaw, C. Identification and characterisation of the antimicrobial peptide, phylloseptin-PT, from the skin secretion of Phyllomedusa tarsius, and comparison of activity with designed, cationicity-enhanced analogues and diastereomers. Molecules 2016, 21, 1667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Samantray, S.; Schumann, W.; Illig, A.-M.; Carballo-Pacheco, M.; Paul, A.; Barz, B.; Strodel, B. Molecular dynamics simulations of protein aggregation: Protocols for simulation setup and analysis with Markov state models and transition networks. bioRxiv 2020, 1–34. [Google Scholar] [CrossRef]










| Peptide | Sequence | Length (Residues) | Mono Molecular Weight (Da) | Charge |
|---|---|---|---|---|
| TPF | FLPLIAGLFGKIF-NH2 | 13 | 1433.88 | 2 |
| des-Phe1 TPF | LPLIAGLFGKIF-NH2 | 12 | 1286.81 | 2 |
| W-des-Phe1 TPF | LPLIAGLWGKIW-NH2 | 12 | 1364.83 | 2 |
| dW-des-Phe1 TPF | LPLIAGLWGKIW-NH2 | 12 | 1364.83 | 2 |
| Peptide | Helix Content % | ||
|---|---|---|---|
| 50% TFE | DPPG/DPPE | DPPC | |
| TPF | 58.9 | 41.4 | 27.7 |
| des-Phe1 TPF | 37.7 | 26.7 | 16.4 |
| W-des-Phe1 TPF | 27.7 | 26.9 | 10.1 |
| dW-des-Phe1 TPF | 3.0 | 0.5 | 8.3 |
| Peptide | MIC/MBC (µM) | |||||
|---|---|---|---|---|---|---|
| S. aureus | MRSA | E. faecalis | E. coli | P. aeruginosa | K. pneumoniae | |
| TPF | 4/32 | 4/4 | 16/16 | >128/>128 | >128/>128 | >128/>128 |
| des-Phe1 TPF | 16/16 | 32/32 | 32/32 | 64/128 | >128/>128 | 128/128 |
| W-des-Phe1 0TPF | 32/32 | 128/128 | 128/128 | 128/128 | >128/>128 | 128/128 |
| dW-des-Phe1 TPF | 64/64 | >128/>128 | >128/>128 | >128/>128 | >128/>128 | >128/>128 |
| Peptide | MBIC/MBEC (µM) | |
|---|---|---|
| S. aureus | MRSA | |
| TPF | 4/32 | 4/16 |
| des-Phe1 TPF | 16/32 | 32/32 |
| W-des-Phe1 TPF | 64/>128 | 128/>128 |
| dW-des-Phe1 TPF | >128/>128 | >128/>128 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zai, Y.; Xi, X.; Ye, Z.; Ma, C.; Zhou, M.; Chen, X.; Siu, S.W.I.; Chen, T.; Wang, L.; Kwok, H.F. Aggregation and Its Influence on the Bioactivities of a Novel Antimicrobial Peptide, Temporin-PF, and Its Analogues. Int. J. Mol. Sci. 2021, 22, 4509. https://doi.org/10.3390/ijms22094509
Zai Y, Xi X, Ye Z, Ma C, Zhou M, Chen X, Siu SWI, Chen T, Wang L, Kwok HF. Aggregation and Its Influence on the Bioactivities of a Novel Antimicrobial Peptide, Temporin-PF, and Its Analogues. International Journal of Molecular Sciences. 2021; 22(9):4509. https://doi.org/10.3390/ijms22094509
Chicago/Turabian StyleZai, Yu, Xinping Xi, Zhuming Ye, Chengbang Ma, Mei Zhou, Xiaoling Chen, Shirley W. I. Siu, Tianbao Chen, Lei Wang, and Hang Fai Kwok. 2021. "Aggregation and Its Influence on the Bioactivities of a Novel Antimicrobial Peptide, Temporin-PF, and Its Analogues" International Journal of Molecular Sciences 22, no. 9: 4509. https://doi.org/10.3390/ijms22094509
APA StyleZai, Y., Xi, X., Ye, Z., Ma, C., Zhou, M., Chen, X., Siu, S. W. I., Chen, T., Wang, L., & Kwok, H. F. (2021). Aggregation and Its Influence on the Bioactivities of a Novel Antimicrobial Peptide, Temporin-PF, and Its Analogues. International Journal of Molecular Sciences, 22(9), 4509. https://doi.org/10.3390/ijms22094509






_Kwok.png)