Effects of G-Quadruplex-Binding Plant Secondary Metabolites on c-MYC Expression
Abstract
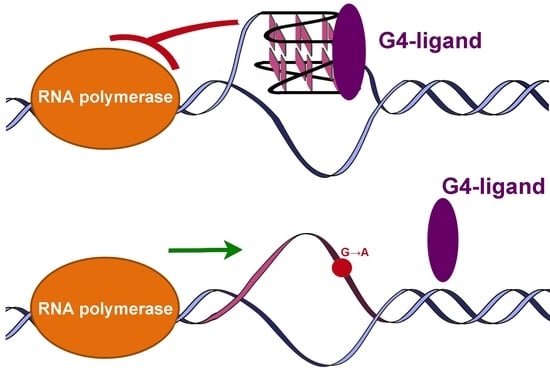
:1. Introduction
2. Results
2.1. Evaluation of PSM Binding to the Promoter G4s Using the G4-FID Assay
2.2. The Thermal Stability of c-MYC G4 Measured by UV and CD Spectroscopy Methods in the Presence of PSMs
2.3. PSMs Down-Regulate the Promoter Activity of c-MYC by Targeting Its G4 Structure
2.4. PSM-Dependent c-MYC Gene Expression in Cultured Cells
3. Discussion
4. Materials and Methods
4.1. Genetal Materials
4.2. Fluorescent Indicator Displacement Assay
4.3. UV Spectroscopy Melting Curves
4.4. CD Measurements
4.5. Cell Cultures
4.6. Cell Viability Assay
4.7. Construction of Reporter Luciferase Vectors, Transfection, and Luciferase Assays
- forward: 5′-CGACCTGGTACCCAGCTGTTCCGCCTGCGATGATTTATAC-3′,
- reverse: 5′-CTCGTCAAGCTTCAGCTGCAAGGAGAGCCTTTC-3′.
- forward: 5′-pGGGGAGGGTGAGGAGGGTGGGGAAGG-3′,
- reverse: 5′-ATAAGCGCCCCTCCCGGGTTC-3′.
- wild-type: 5′-GGGGAGGGTGGGGAGGGTGGGGAAGG-3′,
- mutated: 5′-GGGGAGGGTGAGGAGGGTGGGGAAGG-3′ (substitution underlined).
4.8. RNA Isolation, cDNA Synthesis, and qRT-PCR Assay
- RPL27 (forward): 5′-ACCGTCACCCCCGCAAAGTG-3′,
- RPL27 (reverse): 5′-CCCGTCGGGCCTTGCGTTTA-3′,
- c-MYC (forward): 5′-GGGAGGCTATTCTGCCCATTT-3′,
- c-MYC (reverse): 5′-CGTAGTCGAGGTCATAGTTCCTG-3′,
- RPLP0 (forward): 5′-CCTCGTGGAAGTGACATCGT-3′,
- RPLP0 (reverse): 5′-CTGTCTTCCCTGGGCATCAC-3′,
- GPI (forward): 5′-GGAGACCATCACGAATGCAGA-3′,
- GPI (reverse): 5′-TAGACAGGGCAACAAAGTGCT-3′.
4.9. PCR Array
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- Dolinnaya, N.G.; Ogloblina, A.M.; Yakubovskaya, M.G. Structure, properties, and biological relevance of the DNA and RNA G-quadruplexes: Overview 50 years after their discovery. Biochemistry 2016, 81, 1602–1649. [Google Scholar] [CrossRef] [PubMed]
- Valton, A.-L.; Hassan-Zadeh, V.; Lema, I.; Boggetto, N.; Alberti, P.; Saintomé, C.; Riou, J.-F.; Prioleau, M.-N. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014, 33, 732–746. [Google Scholar] [CrossRef] [PubMed]
- Cheung, I.; Schertzer, M.; Rose, A.; Lansdorp, P.M. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002, 31, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Neidle, S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009, 13, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Zaug, A.J.; Podell, E.R.; Cech, T.R. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 10864–10869. [Google Scholar] [CrossRef]
- Beaume, N.; Pathak, R.; Yadav, V.K.; Kota, S.; Misra, H.S.; Gautam, H.K.; Chowdhury, S. Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: Radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 2013, 41, 76–89. [Google Scholar] [CrossRef]
- Kruisselbrink, E.; Guryev, V.; Brouwer, K.; Pontier, D.B.; Cuppen, E.; Tijsterman, M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr. Biol. 2008, 18, 900–905. [Google Scholar] [CrossRef]
- Halder, R.; Halder, K.; Sharma, P.; Garg, G.; Sengupta, S.; Chowdhury, S. Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol. BioSyst. 2010, 6, 2439–2447. [Google Scholar] [CrossRef]
- Schiavone, D.; Guilbaud, G.; Murat, P.; Papadopoulou, C.; Sarkies, P.; Prioleau, M.-N.; Balasubramanian, S.; Sale, J.E. Determinants of G quadruplex-induced epigenetic instability in REV1-deficient cells. EMBO J. 2014, 33, 2507–2520. [Google Scholar] [CrossRef]
- Lago, S.; Nadai, M.; Cernilogar, F.M.; Kazerani, M.; Moreno, H.D.; Schotta, G.; Richter, S.N. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 2021, 12, 3885. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L. Structure, location and interactions of G-quadruplexes. FEBS J. 2010, 277, 3452–3458. [Google Scholar] [CrossRef]
- Chaudhuri, R.; Bhattacharya, S.; Dash, J.; Bhattacharya, S. Recent Update on Targeting c-MYC G-Quadruplexes by Small Molecules for Anticancer Therapeutics. J. Med. Chem. 2021, 64, 42–70. [Google Scholar] [CrossRef] [PubMed]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent Progress of Targeted G-Quadruplex-Preferred Ligands toward Cancer Therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Chakraborty, S.; Sengupta, P.K.; Bhowmik, S. Exploring the Interactions of the Dietary Plant Flavonoids Fisetin and Naringenin with G-Quadruplex and Duplex DNA, Showing Contrasting Binding Behavior: Spectroscopic and Molecular Modeling Approaches. J. Phys. Chem. B 2016, 120, 8942–8952. [Google Scholar] [CrossRef] [PubMed]
- Salem, A.A.; El Haty, I.A.; Abdou, I.M.; Mu, Y. Interaction of human telomeric G-quadruplex DNA with thymoquinone: A possible mechanism for thymoquinone anticancer effect. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2015, 1850, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Tawani, A.; Mishra, S.K.; Kumar, A. Structural insight for the recognition of G-quadruplex structure at human c-myc promoter sequence by flavonoid Quercetin. Sci. Rep. 2017, 7, 3600. [Google Scholar] [CrossRef]
- Ghosh, S.; Pradhan, S.K.; Kar, A.; Chowdhury, S.; Dasgupta, D. Molecular basis of recognition of quadruplexes human telomere and c-myc promoter by the putative anticancer agent sanguinarine. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2013, 1830, 4189–4201. [Google Scholar] [CrossRef]
- Dickerhoff, J.; Brundridge, N.; McLuckey, S.A.; Yang, D. Berberine Molecular Recognition of the Parallel MYC G-Quadruplex in Solution. J. Med. Chem. 2021, 64, 16205–16212. [Google Scholar] [CrossRef]
- Del Villar-Guerra, R.; Gray, R.D.; Trent, J.O.; Chaires, J.B. A rapid fluorescent indicator displacement assay and principal component/cluster data analysis for determination of ligand-nucleic acid structural selectivity. Nucleic Acids Res. 2018, 46, e41. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. MYC, Metabolism, and Cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Debnath, M.; Müller, D.; Paul, R.; Das, T.; Bessi, I.; Schwalbe, H.; Dash, J. Cell penetrating thiazole peptides inhibit c-MYC expression via site-specific targeting of c-MYC G-quadruplex. Nucleic Acids Res. 2018, 46, 5355–5365. [Google Scholar] [CrossRef]
- Agrawal, P.; Hatzakis, E.; Guo, K.; Carver, M.; Yang, D. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: Insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013, 41, 10584–10592. [Google Scholar] [CrossRef]
- Mathad, R.I.; Hatzakis, E.; Dai, J.; Yang, D. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III1 element: Insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011, 39, 9023–9033. [Google Scholar] [CrossRef]
- Mondal, S.; Jana, J.; Sengupta, P.; Jana, S.; Chatterjee, S. Myricetin arrests human telomeric G-quadruplex structure: A new mechanistic approach as an anticancer agent. Mol. BioSyst. 2016, 12, 2506–2518. [Google Scholar] [CrossRef]
- Bao, H.L.; Liu, H.S.; Xu, Y. Hybrid-type and two-tetrad antiparallel telomere DNA G-quadruplex structures in living human cells. Nucleic Acids Res. 2019, 47, 4940–4947. [Google Scholar] [CrossRef]
- Agrawal, P.; Lin, C.; Mathad, R.I.; Carver, M.; Yang, D. The major G-quadruplex formed in the human BCL-2 proximal promoter adopts a parallel structure with a 13-nt loop in K+ solution. J. Am. Chem. Soc. 2014, 136, 1750–1753. [Google Scholar] [CrossRef]
- Yang, D.; Hurley, L.H. Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides, Nucleotides Nucleic Acids 2006, 25, 951–968. [Google Scholar] [CrossRef]
- Wang, J.M.; Huang, F.C.; Kuo, M.H.; Wang, Z.F.; Tseng, T.Y.; Chang, L.C.; Yen, S.J.; Chang, T.C.; Lin, J.J. Inhibition of cancer cell migration and invasion through suppressing the Wnt1-mediating signal pathway by G-quadruplex structure stabilizers. J. Biol. Chem. 2014, 289, 14612–14623. [Google Scholar] [CrossRef]
- Cogoi, S.; Xodo, L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006, 34, 2536–2549. [Google Scholar] [CrossRef] [PubMed]
- Le, V.H.; Nagesh, N.; Lewis, E.A. Bcl-2 promoter sequence G-quadruplex interactions with three planar and non-planar cationic porphyrins: TMPyP4, TMPyP3, and TMPyP2. PLoS ONE 2013, 8, e72462. [Google Scholar] [CrossRef] [PubMed]
- Amato, J.; Pagano, B.; Borbone, N.; Oliviero, G.; Gabelica, V.; Pauw, E.D.; D’Errico, S.; Piccialli, V.; Varra, M.; Giancola, C.; et al. Targeting G-quadruplex structure in the human c-Kit promoter with short PNA sequences. Bioconj. Chem. 2011, 22, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Monchaud, D.; Allain, C.; Teulade-Fichou, M.P. Development of a fluorescent intercalator displacement assay (G4-FID) for establishing quadruplex-DNA affinity and selectivity of putative ligands. Bioorg. Med. Chem. Lett. 2006, 16, 4842–4845. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.P. G4-FID: A fluorescent DNA probe displacement assay for rapid evaluation of quadruplex ligands. Methods Mol. Biol. 2010, 608, 257–271. [Google Scholar] [CrossRef]
- Ilyinsky, N.S.; Shchyolkina, A.K.; Borisova, O.F.; Mamaeva, O.K.; Zvereva, M.I.; Azhibek, D.M.; Livshits, M.A.; Mitkevich, V.A.; Balzarini, J.; Sinkevich, Y.B.; et al. Novel multi-targeting anthra[2,3-b]thiophene-5,10-diones with guanidine-containing side chains: Interaction with telomeric G-quadruplex, inhibition of telomerase and topoisomerase I and cytotoxic properties. Eur. J. Med. Chem. 2014, 85, 605–614. [Google Scholar] [CrossRef]
- Zenkov, R.G.; Vlasova, O.A.; Maksimova, V.P.; Fetisov, T.I.; Karpechenko, N.Y.; Ektova, L.V.; Eremina, V.A.; Popova, V.G.; Usalka, O.G.; Lesovaya, E.A.; et al. Molecular Mechanisms of Anticancer Activity of N-Glycosides of Indolocarbazoles LCS-1208 and LCS-1269. Molecules 2021, 26, 7329. [Google Scholar] [CrossRef]
- Zidanloo, S.G.; Hosseinzadeh Colagar, A.; Ayatollahi, H.; Raoof, J.B. Downregulation of the WT1 gene expression via TMPyP4 stabilization of promoter G-quadruplexes in leukemia cells. Tumour Biol. 2016, 37, 9967–9977. [Google Scholar] [CrossRef]
- Sengupta, P.; Bhattacharya, A.; Sa, G.; Das, T.; Chatterjee, S. Truncated G-Quadruplex Isomers Cross-Talk with the Transcription Factors To Maintain Homeostatic Equilibria in c-MYC Transcription. Biochemistry 2019, 58, 1975–1991. [Google Scholar] [CrossRef]
- Ragazzon, P.A.; Bradshaw, T.; Matthews, C.; Iley, J.; Missailidis, S. The characterisation of flavone-DNA isoform interactions as a basis for anticancer drug development. Anticancer Res. 2009, 29, 2273–2283. [Google Scholar] [PubMed]
- Tawani, A.; Kumar, A. Structural Insight into the interaction of Flavonoids with Human Telomeric Sequence. Sci. Rep. 2015, 5, 17574. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Sun, H.; Zhou, H.; Xiang, J.; Tang, Y.; Zhao, C. The interaction of telomeric DNA and C-myc22 G-quadruplex with 11 natural alkaloids. Nucleic Acid Ther. 2012, 22, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.N.; Xie, M.X. Spectroscopic investigation of the interaction between G-quadruplex of KRAS promoter sequence and three isoquinoline alkaloids. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 171, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Mikutis, G.; Karaköse, H.; Jaiswal, R.; LeGresley, A.; Islam, T.; Fernandez-Lahore, M.; Kuhnert, N. Phenolic promiscuity in the cell nucleus—Epigallocatechingallate (EGCG) and theaflavin-3,3′-digallate from green and black tea bind to model cell nuclear structures including histone proteins, double stranded DNA and telomeric quadruplex DNA. Food Funct. 2013, 4, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Chakraborty, S.; Chorell, E.; Sengupta, P.K.; Bhowmik, S. Importance of the hydroxyl substituents in the B-ring of plant flavonols on their preferential binding interactions with VEGF G-quadruplex DNA: Multi-spectroscopic and molecular modeling studies. Int. J. Biol. Macromol. 2018, 118, 629–639. [Google Scholar] [CrossRef]
- Paul, S.; Hossain, S.S.; Samanta, A. Interactions between a Bioflavonoid and c-MYC Promoter G-Quadruplex DNA: Ensemble and Single-Molecule Investigations. J. Phys. Chem. B 2019, 123, 2022–2031. [Google Scholar] [CrossRef]
- Pattanayak, R.; Basak, P.; Sen, S.; Bhattacharyya, M. Interaction of KRAS G-quadruplex with natural polyphenols: A spectroscopic analysis with molecular modeling. Int. J. Biol. Macromol. 2016, 89, 228–237. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.P. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008, 6, 627–636. [Google Scholar] [CrossRef]
- Bessi, I.; Bazzicalupi, C.; Richter, C.; Jonker, H.R.; Saxena, K.; Sissi, C.; Chioccioli, M.; Bianco, S.; Bilia, A.R.; Schwalbe, H.; et al. Spectroscopic, molecular modeling, and NMR-spectroscopic investigation of the binding mode of the natural alkaloids berberine and sanguinarine to human telomeric G-quadruplex DNA. ACS Chem. Biol. 2012, 7, 1109–1119. [Google Scholar] [CrossRef]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Hurley, L.H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: Implications for drug targeting and control of gene expression. J. Med. Chem. 2009, 52, 2863–2874. [Google Scholar] [CrossRef] [PubMed]
- Grand, C.L.; Han, H.; Muñoz, R.M.; Weitman, S.; Von Hoff, D.D.; Hurley, L.H.; Bearss, D.J. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002, 1, 565–573. [Google Scholar] [PubMed]
- Ogloblina, A.M.; Khristich, A.N.; Karpechenko, N.Y.; Semina, S.E.; Belitsky, G.A.; Dolinnaya, N.G.; Yakubovskaya, M.G. Multi-targeted effects of G4-aptamers and their antiproliferative activity against cancer cells. Biochimie 2018, 145, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, A.V.; Kubareva, E.A.; Monakhova, M.V.; Zvereva, M.I.; Dolinnaya, N.G. Impact of G-Quadruplexes on the Regulation of Genome Integrity, DNA Damage and Repair. Biomolecules 2021, 11, 1284. [Google Scholar] [CrossRef]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef]





| Sequence Source | Sequence, 5′-3′ | Secondary Structure Formed | Reference |
|---|---|---|---|
| VEGF | GGGGCGGGCCGGGGGCGGGG | Parallel G4 | [29] |
| c-MYC | GGGGAGGGTGGGGAGGGTGGGGAAGG | [30] | |
| WNT1 | GGGGGCCACCGGGCAGGGGGCGGGGG | [31] | |
| KRAS | GGGCGGTGTGGGAAGAGGGAAGAGGGGGAGG | [32] | |
| BCL2 | AGGGGCGGGCGCGGGAGGAAGGGGGCGGGAGCGGGGC | [33] | |
| c-KIT | GGGAGGGCGCTGGGAGGAGGG | [34] | |
| Tel | GGGTTAGGGTTAGGGTTAGGGTTA | Antiparallel/hybrid G4 | [27] |
| Artificial | CAATCGGATCGAATTCGATCCGATTG | dsDNA hairpin | [15] |
| Sequence | Sanguinarine | Quercetin | EGCG | Kaempferol | Thymoquinone | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| DC50 µM | Kb (×106 M−1) | DC50 µM | Kb (×106 M−1) | DC50 µM | Kb (×106 M−1) | DC50 µM | Kb (×106 M−1) | DC50 µM | Kb (×106 M−1) | |
| c-MYC | 0.32 ± 0.02 | 7.76 ± 0.53 | 0.47 ± 0.03 | 5.33 ± 0.36 | 0.85 ± 0.06 | 2.94 ± 0.20 | 2.16 ± 0.15 | 1.16 ± 0.08 | 3.97 ± 0.25 | 0.63 ± 0.04 |
| WNT1 | 1.24 ± 0.17 | 2.83 ± 0.40 | 1.69 ± 0.24 | 2.07 ± 0.30 | 1.27 ± 0.18 | 2.75 ± 0.39 | 1.16 ± 0.16 | 3.03 ± 0.43 | 1.29 ± 0.19 | 2.71 ± 0.39 |
| VEGFA | 4.07 ± 0.57 | 0.86 ± 0.12 | 1.13 ± 0.16 | 3.10 ± 0.44 | 2.00 ± 0.28 | 1.75 ± 0.25 | 1.67 ± 0.24 | 2.09 ± 0.30 | 2.33 ± 0.33 | 1.50 ± 0.21 |
| c-KIT | 1.07 ± 0.15 | 3.28 ± 0.47 | 0.92 ± 0.13 | 3.80 ± 0.54 | 2.43 ± 0.35 | 1.44 ± 0.21 | 2.15 ± 0.30 | 1.63 ± 0.23 | 2.17 ± 0.31 | 1.61 ± 0.23 |
| BCL2 | 0.85 ± 0.12 | 4.12 ± 0.59 | 0.63 ± 0.09 | 5.56 ± 0.79 | 0.96 ± 0.14 | 3.65 ± 0.52 | 2.00 ± 0.28 | 1.75 ± 0.25 | 1.22 ± 0.10 | 2.87 ± 0.41 |
| KRAS | 3.76 ± 0.53 | 0.93 ± 0.13 | 2.03 ± 0.30 | 1.72 ± 0.25 | 2.16 ± 0.31 | 1.62 ± 0.23 | 2.15 ± 0.31 | 1.63 ± 0.23 | 2.30 ± 0.33 | 1.52 ± 0.22 |
| Tel | 8.42 ± 0.59 | 0.43 ± 0.03 | 0.49 ± 0.04 | 7.43 ± 0.55 | 2.00 ± 0.15 | 1.81 ± 0.14 | 2.00 ± 0.15 | 1.81 ± 0.14 | 2.15 ± 0.17 | 1.68 ± 0.13 |
| DNA hairpin | 3.08 ± 0.40 | 1.30 ± 0.17 | 105.26 ± 11.08 | 0.038 ± 0.004 | NA | NA | 114.29 ± 6.53 | 0.035 ± 0.002 | - | 0 |
| Selectivity factor (DC50 DNA hairpin/DC50 c-MYC) | ||||||||||
| c-MYC | 9.63 ± 0.19 | 223.96 ± 0.17 | NA | 52.91 ± 0.13 | Absolute | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zenkov, R.G.; Kirsanov, K.I.; Ogloblina, A.M.; Vlasova, O.A.; Naberezhnov, D.S.; Karpechenko, N.Y.; Fetisov, T.I.; Lesovaya, E.A.; Belitsky, G.A.; Dolinnaya, N.G.; et al. Effects of G-Quadruplex-Binding Plant Secondary Metabolites on c-MYC Expression. Int. J. Mol. Sci. 2022, 23, 9209. https://doi.org/10.3390/ijms23169209
Zenkov RG, Kirsanov KI, Ogloblina AM, Vlasova OA, Naberezhnov DS, Karpechenko NY, Fetisov TI, Lesovaya EA, Belitsky GA, Dolinnaya NG, et al. Effects of G-Quadruplex-Binding Plant Secondary Metabolites on c-MYC Expression. International Journal of Molecular Sciences. 2022; 23(16):9209. https://doi.org/10.3390/ijms23169209
Chicago/Turabian StyleZenkov, Roman G., Kirill I. Kirsanov, Anna M. Ogloblina, Olga A. Vlasova, Denis S. Naberezhnov, Natalia Y. Karpechenko, Timur I. Fetisov, Ekaterina A. Lesovaya, Gennady A. Belitsky, Nina G. Dolinnaya, and et al. 2022. "Effects of G-Quadruplex-Binding Plant Secondary Metabolites on c-MYC Expression" International Journal of Molecular Sciences 23, no. 16: 9209. https://doi.org/10.3390/ijms23169209
APA StyleZenkov, R. G., Kirsanov, K. I., Ogloblina, A. M., Vlasova, O. A., Naberezhnov, D. S., Karpechenko, N. Y., Fetisov, T. I., Lesovaya, E. A., Belitsky, G. A., Dolinnaya, N. G., & Yakubovskaya, M. G. (2022). Effects of G-Quadruplex-Binding Plant Secondary Metabolites on c-MYC Expression. International Journal of Molecular Sciences, 23(16), 9209. https://doi.org/10.3390/ijms23169209