LncGSAR Controls Ovarian Granulosa Cell Steroidogenesis via Sponging MiR-125b to Activate SCAP/SREBP Pathway
Abstract
:1. Introduction
2. Results
2.1. LncRNA-miRNA-mRNA Interaction Network
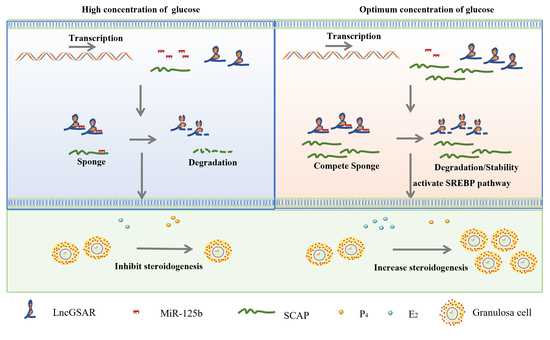
2.2. High Glucose Regulates lncGSAR Expression by Affecting Its Stability
2.3. LncGSAR Promotes Steroid Hormone Secretion and Proliferation of GCs
2.4. LncGSAR Acts as a Molecular Sponge for MiR-125b
2.5. MiR-125b Suppresses GCs Proliferation and Steroidogenesis
2.6. MiR-125b Regulates GCs Proliferation and Steroidogenesis by Targeting SCAP-SREBP Axis
2.7. SCAP Is Involved in GCs Proliferation and Steroidogenesis via Activate SREBP
2.8. LncRNA GSAR Promotes GCs Proliferation and Steroidogenesis via Activating the SCAP-SREBP Pathway by Acting as an MiR-125b Sponge
3. Discussion
4. Materials and Methods
4.1. Cell Collections and Culture
4.2. The Granulosa Cell (GC) Model of Glucose Treatment
4.3. RNA Sequencing (RNA-Seq) and Bioinformatics Analysis
4.4. RNA Extraction, Complementary DNA (cDNA) Synthesis, and (Quantitative Real-Time PCR) qRT-PCR
4.5. Nuclear and Cytoplasmic RNA Fractionation Assay
4.6. Plasmid Construction, RNA Oligonucleotides, and Cell Transfection
4.7. CCK-8 Assay
4.8. Flow Cytometric Analysis
4.9. 5′-bromo-2′-deoxyuridine (BrdU) Detection
4.10. Quantification of E2 and P4 in Culture Medium
4.11. Western Blotting
4.12. Dual-Luciferase Reporter Assay
4.13. In Vitro RNA-RNA Interaction Pull-Down Assay
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Jahromi, B.N.; Mosallanezhad, Z.; Matloob, N.; Davari, M.; Ghobadifar, M.A. The potential role of granulosa cells in the maturation rate of immature human oocytes and embryo development: A co-culture study. Clin. Exp. Reprod. Med. 2015, 42, 111–117. [Google Scholar] [CrossRef] [PubMed]
- McGee, E.A.; Hsueh, A.J. Initial and cyclic recruitment of ovarian follicles. Endocr. Rev. 2000, 21, 200–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumesic, D.A.; Meldrum, D.R.; Katz-Jaffe, M.G.; Krisher, R.L.; Schoolcraft, W.B. Oocyte environment: Follicular fluid and cumulus cells are critical for oocyte health. Fertil. Steril. 2015, 103, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Charlier, C.; Montfort, J.; Chabrol, O.; Brisard, D.; Nguyen, T.; Aurélie, L.C.; Laurent, R.P.; François, M.; Pierre, P.; Svetlana, U.; et al. Oocyte-somatic cells interactions, lessons from evolution. BMC Genom. 2012, 13, 560. [Google Scholar] [CrossRef] [Green Version]
- Fraunhoffer, N.A.; Abuelafia, M.A.; Barrientos, M.A.; Cimerman, K.V.; Olmos, M.F.; Chuluyan, E.; Barrios, M. Long-term apoptosis-related protein expression in the diabetic mouse ovary. PLoS ONE 2018, 13, e0203268. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Duan, C.H.; Guo, Y.X.; Li, J.J.; He, H.Y.; Li, R.T.; Liu, Y.Q.; Zhang, Y.J. Effects of glucose on glycolysis and steroidogenesis as well as related gene expression in ovine granulosa cells in vitro. Small Rumin. Res. 2022, 215, 106766. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, Y.X.; Duan, C.H.; Yang, R.C.; Zhang, L.C.; Liu, Y.Q.; Zhang, Y.J. Long Non-Coding RNA GDAR Regulates Ovine Granulosa Cells Apoptosis by Affecting the Expression of Apoptosis-Related Genes. Int. J. Mol. Sci. 2022, 23, 5183. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Zou, X.; Lu, T.; Deng, M.; Li, Y.; Guo, Y.; Sun, B.L.; Liu, G.B.; Liu, D.W. Identification of mRNAs and lncRNAs Involved in the Regulation of Follicle Development in Goat. Front. Genet. 2020, 11, 589076. [Google Scholar] [CrossRef]
- Wang, J.; Niu, M.; Zhang, T.; Shen, W.; Cao, H. Genome-Wide Network of lncRNA–mRNA During Ovine Oocyte Development from Germinal Vesicle to Metaphase II in vitro. Front. Physiol. 2020, 11, 1019. [Google Scholar] [CrossRef]
- Luo, J.; Wang, K.; Yeh, S.; Sun, Y.; Liang, L.; Xiao, Y.; Xu, W.H.; Niu, Y.J.; Cheng, L.; Sankar, N.M.; et al. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat. Commun. 2019, 10, 2571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.Y.; Zhao, J.; Geng, X.Y.; Chu, W.W.; Li, S.; Chen, Z.J.; Du, Y.Z. Long non-coding RNA lnc-CCNL1-3:1 promotes granulosa cell apoptosis and suppresses glucose uptake in women with polycystic ovary syndrome. Mol. Ther. Nucleic Acid. 2020, 23, 614–628. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.L.; Liu, S.X.; Xing, G.L.; Wang, F. lncRNA PVT1/MicroRNA-17-5p/PTEN Axis Regulates Secretion of E2 and P4, Proliferation, and Apoptosis of Ovarian Granulosa Cells in PCOS. Mol. Ther. Nucleic Acid. 2020, 20, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Zhen, J.R.; Li, J.L.; Li, X.; Wang, X.; Xiao, Y.L.; Sun, Z.Y.; Yu, Q. Downregulating lncRNA NEAT1 induces proliferation and represses apoptosis of ovarian granulosa cells in polycystic ovary syndrome via microRNA-381/IGF1 axis. J. Biomed. Sci. 2021, 28, 53. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.L.; Gao, X.X.; Bao, Y.J.; El-Samahy, M.A.; Yang, J.Y.; Wang, Z.B.; Li, X.D.; Zhang, G.M.; Zhang, Y.L.; Liu, W.J.; et al. lncRNA FDNCR promotes apoptosis of granulosa cells by targeting the miR-543-3p/DCN/TGF-b signaling pathway in Hu sheep. Mol. Ther. Nucleic Acid. 2021, 24, 223–240. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, Z.X.; Du, X.; Zhang, J.B.; Liu, H.L.; Li, Q.F. NORHA, a novel follicular atresia-related lncRNA, promotes porcine granulosa cell apoptosis via the miR-183-96-182 cluster and FoxO1 axis. J. Anim. Sci. Biotechnol. 2021, 12, 103. [Google Scholar] [CrossRef]
- Cheng, C.M.; Geng, F.; Li, Z.; Zhong, Y.G.; Wang, H.B.; Cheng, X.; Zhao, Y.; Mo, X.K.; Horbinski, C.; Duan, W.R.; et al. Ammonia stimulates SCAP/Insig dissociation and SREBP-1 activation to promote lipogenesis and tumour growth. Nat. Metab. 2022, 4, 575–588. [Google Scholar] [CrossRef]
- Shimizu-Albergine, M.; Yserloo, V.B.; Golkowski, M.G.; Ong, S.E.; Beavo, J.A.; Bornfeldt, K.E. SCAP/SREBP pathway is required for the full steroidogenic response to cyclic AMP. Proc. Natl. Acad. Sci. USA 2016, 113, E5685–E5693. [Google Scholar] [CrossRef] [Green Version]
- Song, P.Y.; Yue, Q.X.; Fu, Q.; Li, X.Y.; Li, X.J.; Zhou, R.Y.; Chen, X.R.; Tao, C.Y. Integrated Analysis of miRNA-mRNA interaction in ovaries of Turpan Black Sheep during follicular and luteal phases. Reprod. Domest. Anim. 2020, 56, 46–57. [Google Scholar] [CrossRef]
- Yao, Y.L.; Niu, J.Q.; Sizhu, S.L.; Li, B.J.; Chen, Y.; Li, R.Y.; Yangzong, Q.B.; Li, Q.F.; Xu, Y.F. microRNA-125b Regulates Apoptosis by Targeting Bone Morphogenetic Protein Receptor 1B in Yak Granulosa Cells. DNA Cell Biol. 2018, 37, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Sun, L.; Feramisco, J.D.; Brown, M.S.; Goldstein, J.L. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell 2002, 10, 237–245. [Google Scholar] [CrossRef]
- Walsh, S.W.; Matthews, D.; Browne, J.A.; Forde, N.; Crowe, M.A.; Mihm, M.; Diskin, M.; Evans, A.C.O. Acute dietary restriction in heifers alters expression of genes regulating exposure and response to gonadotrophins and IGF in dominant follicles. Anim. Reprod. Sci. 2012, 133, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Comin, A.; Peric, T.; Montillo, M.; Cappa, A.; Marchi, V.; Veronesi, M.C.; Prandi, A. Luteal activity and effect of dietary energy restriction on follicular development in lactating cows. Reprod. Domest. Anim. 2017, 52, 632–639. [Google Scholar] [CrossRef]
- Ying, S.J.; Wang, Z.Y.; Wang, C.L.; Nie, H.T.; He, D.Y.; Jia, R.X.; Wu, Y.C.; Wan, Y.J.; Zhou, Z.R.; Yan, Y.B.; et al. Effect of different levels of short-term feed intake on folliculogenesis and follicular fluid and plasma concentrations of lactate dehydrogenase, glucose, and hormones in Hu sheep during the luteal phase. Reproduction 2011, 142, 699–710. [Google Scholar] [CrossRef] [Green Version]
- Brownlee, M. Biochemicstry and molecular cell biology of diabetic complications. Nature 2001, 414, 813–820. [Google Scholar] [CrossRef]
- Mauvais-Jarvis, F.; Clegg, D.J.; Hevener, A.L. The role of estrogens in control of energy balanceand glucose homeostasis. Endocr. Rev. 2013, 34, 309–338. [Google Scholar] [CrossRef] [Green Version]
- El-Bahr, S.M.; Ghoneim, I.M.; Waheed, M.M. Biochemical and hormonal analysis of follicular fluid and serum of female dromedary camels (Camelus dromedarius) with different sized ovarian follicles. Anim. Reprod. Sci. 2015, 159, 98–103. [Google Scholar] [CrossRef]
- Guo, Y.X.; Duan, C.H.; Hao, Q.H.; Liu, Y.Q.; Li, T.; Zhang, Y.J. Effect of short-term nutritional supplementation on hormone concentrations in ovarian follicular fluid and steroid regulating gene mRNA abundances in granulosa cells of ewes. Anim. Reprod. Sci. 2019, 211, 106208. [Google Scholar] [CrossRef]
- Li, X.L.; Zhang, T.Y.; Li, S.X.; Deng, Y.Y.; Wang, L.H.; Tao, T.; Wang, S.J.; Gu, Y.Y.; Gu, W.Q.; Hong, J.; et al. Correlation between glucose metabolism and serum steroid hormones in patients with polycystic ovary syndrome. Clin. Endocrinol. 2020, 92, 350–357. [Google Scholar] [CrossRef]
- Chabrolle, C.; JeanPierre, E.; Tosca, L.; Ramé, C.; Dupont, J. Effects of high levels of glucose on the steroidogenesis and the expression of adiponectin receptors in rat ovarian cells. Reprod. Biol. Endocrinol. 2008, 6, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, J.; Shi, B.; Wang, T.R.; Fang, Y.Y.; Cao, T.F.; Zhou, Y.M.; Wang, X.X.; Li, D. Characterization of long non-coding RNA and messenger RNA profiles in follicular fluid from mature and immature ovarian follicles of healthy women and women with polycystic ovary syndrome. Hum. Reprod. 2018, 33, 1735–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, W.Y.; Ding, B.S.; Zhong, G.S.; Du, C.Y.; Fan, W.M.; Fu, P.F. Dysregulation of pseudogene/lncRNA-hsa-miR-363-3p-SPOCK2 pathway fuels stage progression of ovarian cancer. Aging 2019, 11, 11416–11439. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.J.; Zhang, P.; Li, Y.M.; Zhao, N.; Han, H.Y. The AMPK-mTOR axis requires increased MALAT1 expression for promoting granulosa cell proliferation in endometriosis. Exp. Ther. Med. 2021, 21, 21. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. Emerging roles for natural microRNA sponges. Curr. Biol. 2010, 20, R858–R861. [Google Scholar] [CrossRef] [Green Version]
- Karreth, F.A.; Pandolfi, P.P. ceRNA cross-talk in cancer: When ce-bling rivalries go awry. Cancer Discov. 2013, 3, 1113–1121. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Li, H.; Li, P.Y.; Li, X.; Lin, J.J.; Zhu, L.L.; Silva, M.A.; Wang, X.F.; Wang, P.; Zhang, Z. Long noncoding RNA MLK7-AS1 promotes ovarian cancer cells progression by modulating miR-375/YAP1 axis. J. Exp. Clin. Cancer Res. 2018, 37, 237. [Google Scholar] [CrossRef]
- Jiang, X.Z.; Ye, Z.X.; Jiang, Y.F.; Yu, W.; Fang, Q. LncRNA OIP5-AS1 upregulates snail expression by sponging miR-34a to promote ovarian carcinoma cell invasion and migration. Biol. Res. 2020, 53, 49. [Google Scholar] [CrossRef]
- Wang, X.M.; Meng, K.; Wang, H.Q.; Wang, Y.; Zhao, Y.Q.; Kang, J.; Zhang, Y.; Quan, F.S. Identification of small extracellular vesicle subtypes in follicular fluid: Insights into the function and miRNA profiles. J. Cell. Physiol. 2021, 236, 5633–5645. [Google Scholar] [CrossRef]
- Guo, Y.; Wu, Y.B.; Shi, J.H.; Zhuang, H.; Ci, L.; Huang, Q.; Wan, Z.P.; Yang, H.; Zhang, M.J.; Tan, Y.T.; et al. miR-29a/b 1 Regulates the Luteinizing Hormone Secretion and Affects Mouse Ovulation. Front. Endocrinol. 2021, 12, 636220. [Google Scholar] [CrossRef]
- Li, G.X.; Chen, Y.; Jin, W.J.; Zhai, B.; Li, Y.F.; Sun, G.R.; Li, H.; Kang, X.T.; Tian, Y.D. Effects of MiR-125b-5p on Preadipocyte Proliferation and Differentiation in Chicken. Mol. Biol. Rep. 2021, 48, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.Z.; Li, G.P.; Wang, C.G.; Gong, G.Q.; Wang, L.W.; Li, C.Y.; Chen, Y.; Wang, X.L. MIR210HG regulates glycolysis, cell proliferation, and metastasis of pancreatic cancer cells through MiR-125b-5p/HK2/PKM2 axis. RNA Biol. 2021, 18, 2513–2530. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Prizant, H.; Light, A.; Biswas, A.; Hayes, E.; Lee, H.J.; Barad, D.; Gleicher, N.; Hammes, S.R. Androgens regulate ovarian follicular development by increasing follicle stimulating hormone receptor and microRNA-125b expression. Proc. Natl. Acad. Sci. USA 2014, 111, 8–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeBose-Boyd, R.A.; Brown, M.S.; Li, W.P.; Nohturfft, A.; Goldstein, J.L.; Espenshade, P.J. Transport-dependent proteolysis of SREBP: Relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 1999, 99, 703–712. [Google Scholar] [CrossRef] [Green Version]
- Espenshade, P.J.; Cheng, D.; Goldstein, J.L.; Brown, M.S. Autocatalytic processing of site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 1999, 274, 22795–22804. [Google Scholar] [CrossRef] [Green Version]
- Espenshade, P.J.; Li, W.P.; Yabe, D. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc. Natl. Acad. Sci. USA 2002, 99, 11694–11699. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Ma, A.T.; Zhang, Y.J.; Liu, Y.Q.; Wang, S.Y.; Shi, X.F.; Gao, K.; Duan, C.H. Effects of silencing INHα gene by RNAi on the mRNA expressions of StAR, FST, INHβB, and FSHR genes in cultured sheep granulosa cells. Small Rumin. Res. 2017, 157, 23–26. [Google Scholar] [CrossRef]
- Somchit, A.; Campbell, B.K.; Khalid, M.; Kendall, N.R.; Scaramuzzi, R.J. The effect of short-term nutritional supplementation of ewes with lupin grain (Lupinus luteus), during the luteal phase of the estrous cycle on the number of ovarian follicles and the concentrations of hormones and glucose in plasma and follicular fluid. Theriogenology 2007, 68, 1037–1046. [Google Scholar] [CrossRef]
- Scaramuzzi, R.J.; Campbell, B.K.; Souza, C.J.H.; Baird, D.T. Glucose uptake and lactate production by the autotransplanted ovary of the ewe during the luteal and follicular phases of the oestrous cycle. Theriogenology 2010, 73, 1061–1067. [Google Scholar] [CrossRef]
- Campbell, B.K.; Onions, V.; Kendall, N.R.; Guo, L.; Scaramuzzi, R.J. The effect of monosaccharide sugars and pyruvate on the differentiation and metabolism of sheep granulosa cells in vitro. Reproduction 2010, 140, 541–550. [Google Scholar] [CrossRef]
- Langhout, D.J.; Spicer, L.J.; Geisert, R.D. Development of a culture system for bovine granulosa cells: Effects of growth hormone, estradiol, and gonadotropins on cell proliferation, steroidogenesis, and protein synthesis. J. Anim. Sci. 1991, 69, 3321–3334. [Google Scholar] [CrossRef] [PubMed]
- Spicer, L.J.; Chamberlain, C.S. Influence of Cortisol on Insulin and Insulin-Like Growth Factor 1 (IGF-1)-Induced Steroid Production and on IGF-1 Receptors in Cultured Bovine Granulosa Cells and Thecal Cells. Endocrine 1998, 9, 153–162. [Google Scholar] [CrossRef]
- Spicer, L.J.; Chamberlain, C.S.; Maciel, S.M. Influence of gonadotropins on insulin and insulin-like growth factor-I (IGF-I)-induced steroid production by bovine granulosa cells. Domest. Anim. Endocrinol. 2002, 22, 237–254. [Google Scholar] [CrossRef]
- Spicer, L.J. Growth differentiation factor-9 has divergent effects on proliferation and steroidogenesis of bovine granulosa cells. J. Endocrinol. 2006, 189, 329–339. [Google Scholar] [CrossRef] [Green Version]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Method. 2012, 9, 1069–1076. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Guo, Y.; Duan, C.; Li, J.; Ji, S.; Yan, H.; Liu, Y.; Zhang, Y. LncGSAR Controls Ovarian Granulosa Cell Steroidogenesis via Sponging MiR-125b to Activate SCAP/SREBP Pathway. Int. J. Mol. Sci. 2022, 23, 12132. https://doi.org/10.3390/ijms232012132
Wang Y, Guo Y, Duan C, Li J, Ji S, Yan H, Liu Y, Zhang Y. LncGSAR Controls Ovarian Granulosa Cell Steroidogenesis via Sponging MiR-125b to Activate SCAP/SREBP Pathway. International Journal of Molecular Sciences. 2022; 23(20):12132. https://doi.org/10.3390/ijms232012132
Chicago/Turabian StyleWang, Yong, Yunxia Guo, Chunhui Duan, Junjie Li, Shoukun Ji, Huihui Yan, Yueqin Liu, and Yingjie Zhang. 2022. "LncGSAR Controls Ovarian Granulosa Cell Steroidogenesis via Sponging MiR-125b to Activate SCAP/SREBP Pathway" International Journal of Molecular Sciences 23, no. 20: 12132. https://doi.org/10.3390/ijms232012132
APA StyleWang, Y., Guo, Y., Duan, C., Li, J., Ji, S., Yan, H., Liu, Y., & Zhang, Y. (2022). LncGSAR Controls Ovarian Granulosa Cell Steroidogenesis via Sponging MiR-125b to Activate SCAP/SREBP Pathway. International Journal of Molecular Sciences, 23(20), 12132. https://doi.org/10.3390/ijms232012132