Alu Deletions in LAMA2 and CDH4 Genes Are Key Components of Polygenic Predictors of Longevity
Abstract
:1. Introduction
2. Results
2.1. Population Analysis
2.2. Age-Dependent Analysis of Individual Alu Polymorphic Loci
2.3. Polygenic Analysis of Longevity
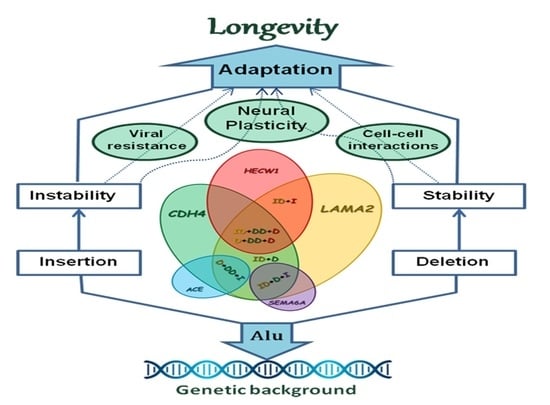
3. Discussion
4. Materials and Methods
4.1. Study Group
4.2. DNA Collection
4.3. Genotyping
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sebastiani, P.; Perls, T.T. The genetics of extreme longevity: Lessons from the new England centenarian study. Front. Genet. 2012, 3, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, Y.I.; Nie, C.; Min, J.; Liu, X.; Li, M.; Chen, H.; Xu, H.; Wang, M.; Ni, T.; Li, Y.; et al. Novel loci and pathways significantly associated with longevity. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Pilling, L.C.; Kuo, C.L.; Sicinski, K.; Tamosauskaite, J.; Kuchel, G.A.; Harries, L.W.; Herd, P.; Wallace, R.; Ferrucci, L.; Melzer, D. Human longevity: 25 genetic loci associated in 389,166 UK biobank participants. Aging 2017, 9, 2504–2520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deelen, J.; Evans, D.S.; Arking, D.E.; Tesi, N.; Nygaard, M.; Liu, X.; Wojczynski, M.K.; Biggs, M.L.; van der Spek, A.; Atzmon, G.; et al. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Gladyshev, V.N. The origin of aging-imperfectness-drive non-random damage defines the aging process and control of lifespan. Trends Genet. 2013, 29, 506–512. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Cho, C.S.; Han, K.; Lee, J. Structural Variation of Alu Element and Human Disease. Genom. Inf. 2016, 14, 70–77. [Google Scholar] [CrossRef] [Green Version]
- Hueso, M.; Cruzado, J.M.; Torras, J.; Navarro, E. ALUminating the Path of Atherosclerosis Progression: Chaos Theory Suggests a Role for Alu Repeats in the Development of Atherosclerotic Vascular Disease. Int. J. Mol. Sci 2018, 19, 1734. [Google Scholar] [CrossRef] [Green Version]
- Kubiak, M.R.; Makałowska, I. Protein-Coding Genes' Retrocopies and Their Functions. Viruses 2017, 9, 80. [Google Scholar] [CrossRef] [Green Version]
- Todd, C.D.; Deniz, Ö.; Taylor, D.; Branco, M.R. Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. Elife 2019, 8, e44344. [Google Scholar] [CrossRef]
- Ule, J. Alu elements: At the crossroads between disease and evolution. Biochem. Soc. Trans. 2013, 41, 1532–1535. [Google Scholar] [CrossRef]
- Klawitter, S.; Fuchs, N.V.; Upton, K.R.; Munoz-Lopez, M.; Shukla, R.; Wang, J.; Garcia-Canadas, M.; Lopez-Ruiz, C.; Gerhardt, D.J.; Sebe, A.; et al. Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedoroff, N.V. Transposable elements, epigenetics, and genome evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Schifanella, L.; Larsen, P.A. Alu retrotransposons and COVID-19 susceptibility and morbidity. Hum. Genom. 2021, 15, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, P.H. What might retrotransposons teach us about aging? Curr. Genet. 2016, 62, 277–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, T.; Lin, Z.; Mai, P.; Zhang, E.; Peng, L. Identification of prognostic biomarkers associated with the occurrence of portal vein tumor thrombus in hepatocellular carcinoma. Aging 2021, 13, 11786–11807. [Google Scholar] [CrossRef]
- Zhou, X.; Huang, H.; Cui, W.; Wang, Y.; Luo, W.; Matskova, L.; Zhou, X. Expression and Prognostic Significance of Cadherin 4 (CDH4) in Renal Cell Carcinoma. Med. Sci. Monit. 2020, 26, e922836. [Google Scholar] [CrossRef]
- Xu, M.; Liu, C.; Pu, L.; Lai, J.; Li, J.; Ning, Q.; Liu, X.; Deng, S. Systemic analysis of the expression levels and prognosis of breast cancer-related cadherins. Exp. Biol. Med. 2021, 246, 1706–1720. [Google Scholar] [CrossRef]
- Du, C.; Huang, T.; Sun, D.; Mo, Y.; Feng, H.; Zhou, X.; Xiao, X.; Yu, N.; Hou, B.; Huang, G.; et al. CDH4 as a novel putative tumor suppressor gene epigenetically silenced by promoter hypermethylation in nasopharyngeal carcinoma. Cancer Lett. 2011; 309, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Ning, G.; Si, P.; Zhang, C.; Liu, W.; Ge, W.; Cui, K.; Zhang, R.; Ge, S. E3 ubiquitin ligase HECW1 promotes the metastasis of non-small cell lung cancer cells through mediating the ubiquitination of Smad4. Biochem. Cell Biol. 2021, 99, 675–681. [Google Scholar] [CrossRef]
- Wang, C.; Dong, K.; Wang, Y.; Peng, G.; Song, X.; Yu, Y.; Cui, X. Integrating HECW1 expression into the clinical indicators exhibits high accuracy in assessing the prognosis of patients with clear cell renal cell carcinoma. BMC Cancer 2021, 21, 890. [Google Scholar] [CrossRef]
- Shen, C.Y.; Chang, Y.C.; Chen, L.H.; Lin, W.C.; Lee, Y.H.; Yeh, S.T.; Chen, H.K.; Fang, W.; Hsu, C.P.; Lee, J.M.; et al. The extracellular SEMA domain attenuates intracellular apoptotic signaling of semaphorin 6A in lung cancer cells. Oncogenesis 2018, 7, 95. [Google Scholar] [CrossRef] [Green Version]
- Seshadri, S.; DeStefano, A.L.; Au, R.; Massaro, J.M.; Beiser, A.S.; Kelly-Hayes, M.; Kase, C.S.; D’Agostino, R.B.; DeCarli, C.; Atwood, L.D.; et al. Genetic correlates of brain aging on MRI and cognitive test measures: A genome-wide association and linkage analysis in the Framingham Study. BMC Med. Genet. 2007, 8, S15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.K.; Au, A.; Menon, S.; Griffiths, L.R.; Kooi, C.W.; Irene, L.; Zhao, J.; Lee, C.; Avdonina, M.A.; Hassan, M.R.A.; et al. Polymorphisms of MTHFR, eNOS, ACE, AGT, ApoE, PON1, PDE4D, and Ischemic Stroke: Meta-Analysis. J. Stroke Cereb. Dis. 2017, 26, 2482–2493. [Google Scholar] [CrossRef] [PubMed]
- Amara, A.; Mrad, M.; Sayeh, A.; Lahideb, D.; Layouni, S.; Haggui, A.; Fekih-Mrissa, N.; Haouala, H.; Nsiri, B. The effect of ACE I/D polymorphisms alone and with concomitant risk factors on coronary artery disease. Clin. Appl. Thromb. Hemost. 2018, 24, 157–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, D.; Brown, L.; Malik, K.; Murakami, S. Two Opposing Functions of Angiotensin-Converting Enzyme (ACE) That Links Hypertension, Dementia, and Aging. Int. J. Mol. Sci. 2021, 22, 13178. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.S.; Hsu, L.J.; Chang, N.S. Functional role of WW domain-containing proteins in tumor biology and diseases: Insight into the role in ubiquitin-proteasome system. FASEB Bioadv. 2020, 2, 234–253. [Google Scholar] [CrossRef] [PubMed]
- Urbich, C.; Kaluza, D.; Frömel, T.; Knau, A.; Bennewitz, K.; Boon, R.A.; Bonauer, A.; Doebene, C.; Boeckel, J.N.; Hergenreider, E.; et al. MicroRNA-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood J. Am. Soc. Hematol. 2012, 119, 1607–1616. [Google Scholar] [CrossRef] [Green Version]
- Lan, X.; Zhang, H.; Wang, Z.; Dong, W.; Sun, W.; Shao, L.; Zhang, T.; Zhang, D. Genome-wide analysis of long noncoding RNA expression profile in papillary thyroid carcinoma. Gene 2015, 569, 109–117. [Google Scholar] [CrossRef]
- Wang, S.; Chu, F.; Xia, R.; Guan, J.; Zhou, L.; Fang, X.; Dai, T.; Xie, F.; Zhang, L.; Zhou, F. LPA Maintains Innate Antiviral Immunity in a Pro-Active State via STK38l-Mediated IRF3 Ser303 Phosphorylation. SSRN Electron. J. 2022. [Google Scholar] [CrossRef]
- Cacquevel, M.; Launay, S.; Castel, H.; Benchenane, K.; Chéenne, S.; Buée, L.; Moons, L.; Delacourte, A.; Carmeliet, P.; Vivien, D. Ageing and amyloid-beta peptide deposition contribute to an impaired brain tissue plasminogen activator activity by different mechanisms. Neurobiol. Dis. 2007, 27, 164–173. [Google Scholar] [CrossRef]
- Garatachea, N.; Marín, P.J.; Lucia, A. The ACE DD genotype and D-allele are associated with exceptional longevity: A meta-analysis. Ageing Res. Rev. 2013, 12, 1079–1087. [Google Scholar] [CrossRef]
- Said, R.; Jenni, R.; Boussetta, S.; Ammous, F.; Zouari, S.; Zaghbib, S.; Chakroun, M.; Derouiche, A.; Chebil, M.; Ouerhani, S. Association of a common genetic variant (insertion/deletion) in ACE gene with prostate cancer susceptibility in a Tunisian population. J. Clin. Lab. Anal. 2022, 36, e24129. [Google Scholar] [CrossRef]
- Yashin, A.I.; Wu, D.; Arbeev, K.G.; Ukraintseva, S.V. Polygenic effects of common single-nucleotide polymorphisms on life span: When association meets causality. Rejuvenation Res. 2012, 15, 381–394. [Google Scholar] [CrossRef] [Green Version]
- Schächter, F.; Faure-Delanef, L.; Guénot, F.; Rouger, H.; Froguel, P.; Lesueur-Ginot, L.; Cohen, D. Genetic associations with human longevity at the APOE and ACE loci. Nat. Genet. 1994, 6, 29–32. [Google Scholar] [CrossRef]
- Kolovou, G.; Kolovou, V.; Vasiliadis, I.; Giannakopoulou, V.; Mihas, C.; Bilianou, H.; Kollia, A.; Papadopoulou, E.; Marvaki, A.; Goumas, G.; et al. The Frequency of 4 Common Gene Polymorphisms in Nonagenarians, Centenarians, and Average Life Span Individuals. Angiology 2014, 65, 210–215. [Google Scholar] [CrossRef]
- Atella, V.; Piano Mortari, A.; Kopinska, J.; Belotti, F.; Lapi, F.; Cricelli, C.; Fontana, L. Trends in age-related disease burden and healthcare utilization. Aging Cell 2019, 18, e12861. [Google Scholar] [CrossRef]
- Chen, L.; He, C.Y.; Su, X.; Peng, J.L.; Chen, D.L.; Ye, Z.; Yang, D.D.; Wang, Z.X.; Wang, F.; Shao, J.Y.; et al. SPP1 rs4754 and its epistatic interactions with SPARC polymorphisms in gastric cancer susceptibility. Gene 2018, 640, 43–50. [Google Scholar] [CrossRef]
- Durbeej, M. Laminins. Cell Tissue Res 2010, 339, 259–268. [Google Scholar] [CrossRef]
- Tunggal, P.; Smyth, N.; Paulsson, M.; Ott, M.C. Laminins: Structure and genetic regulation. Microsc. Res. Tech. 2000, 51, 214–227. [Google Scholar] [CrossRef]
- Miner, J.H. Laminins and their roles on mammals. Microsc. Res. Tech. 2008, 71, 349–356. [Google Scholar] [CrossRef]
- Oliveira, J.; Gruber, A.; Cardoso, M.; Taipa, R.; Fineza, I.; Goncalves, A.; Laner, A.; Winder, T.; Schroeder, J.; Rath, J.; et al. LAMA2 gene mutation update: Toward a more comprehensive picture of the laminin-alpha2 variome and its related phenotypes. Hum. Mutat. 2018, 39, 1314–1337. [Google Scholar] [CrossRef]
- Miyagoe-Suzuki, Y.; Nakagawa, Y.; Takeda, S.I. Merosin and congenital muscular dystrophy. Microsc. Res. Tech. 2000, 48, 181–191. [Google Scholar] [CrossRef]
- Chun, S.J.; Rasband, M.N.; Sidman, R.L.; Habib, A.A.; Vartanian, T. Integrin-linked kinase is required for laminin-2-induced oligodendrocyte cell spreading and CNS myelination. J. Cell Biol. 2003, 163, 397–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, C.; Inestrosa, N.C. Interactions of laminin with the amyloid beta peptide. Implications for Alzheimer's disease. Braz. J. Med. Biol. Res. 2001, 34, 597–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De La Fuente, A.G.; Lange, S.; Silva, M.E.; Gonzalez, G.A.; Tempfer, H.; van Wijngaarden, P.; Zhao, C.; Di Canio, L.; Trost, A.; Bieler, L.; et al. Pericytes Stimulate Oligodendrocyte Progenitor Cell Differentiation during CNS Remyelination. Cell Rep. 2017, 20, 1755–1764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hibbitts, A.J.; Kočí, Z.; Kneafsey, S.; Matsiko, A.; Žilić, L.; Dervan, A.; Hinton, P.; Chen, G.; Cavanagh, B.; Dowling, J.K.; et al. Multi-Factorial Nerve Guidance Conduit Engineering Improves Outcomes in Inflammation, Angiogenesis and Large Defect Nerve Repair. Matrix Biol. 2022, 106, 34–57. [Google Scholar] [CrossRef]
- Kittur, S.D.; Adler, W.H.; Martin, G.R.; Schapiro, M.B.; Rapoport, S.I.; Gunzler, V. Laminin concentrations in serum and cerebrospinal fluid in aging and Alzheimer's disease. Int. J. Dev. Neurosci. 1993, 11, 95–99. [Google Scholar] [CrossRef]
- Hawkes, C.A.; Hartig, W.; Kacza, J.; Schliebs, R.; Weller, R.O.; Nicoll, J.A.; Carare, R.O. Perivascular drainage of solutes is impaired in the ageing mouse brain and in the presence of cerebral amyloid angiopathy. Acta Neuropathol. 2011, 121, 431–443. [Google Scholar] [CrossRef]
- Reed, M.J.; Damodarasamy, M.; Banks, W.A. The extracellular matrix of the blood–brain barrier: Structural and functional roles in health, aging, and Alzheimer’s disease. Tissue Barriers 2019, 7, 1651157. [Google Scholar] [CrossRef]
- Komkov, A.Y.; Maschan, M.A.; Shvets, V.I.; Lebedev, Y.B. Functional analysis of polymorphic insertions of Alu retroelements in acute lymphoblastic leukemia patients. Russ. J. Bioorg. Chem. 2012, 38, 306–318. [Google Scholar] [CrossRef]
- Halbleib, J.M.; Nelson, W.J. Cadherins in development: Cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006, 20, 3199–3214. [Google Scholar] [CrossRef] [Green Version]
- Shan, W.S.; Tanaka, H.; Phillips, G.R.; Arndt, K.; Yoshida, M.; Colman, D.R.; Shapiro, L. Functional cis-heterodimers of N-and R-cadherins. J. Cell Biol. 2000, 148, 579–590. [Google Scholar] [CrossRef]
- Ramirez, T.A.; Saux, J.L.; Joy, A.; Zhang, J.; Dai, Q.; Mifflin, S.; Lindsey, M.L. Chronic and intermittent hypoxia differentially regulate left ventricular inflammatory and extracellular matrix responses. Hypertens. Res. 2012, 35, 811–818. [Google Scholar] [CrossRef] [Green Version]
- Miyazaki, K.; Fujita, T.; Ozaki, T.; Kato, C.; Kurose, Y.; Sakamoto, M.; Kato, S.; Goto, T.; Itoyama, Y.; Aoki, M.; et al. NEDL1, a novel ubiquitin-protein isopeptide ligase for dishevelled-1, targets mutant superoxide dismutase-1. J. Biol. Chem. 2004, 279, 11327–11335. [Google Scholar] [CrossRef] [Green Version]
- Haouari, S.; Vourc’h, P.; Jeanne, M.; Marouillat, S.; Veyrat-Durebex, C.; Lanznaster, D.; Laumonnier, F.; Corcia, P.; Blasco, H.; Andres, C.R. The Roles of NEDD4 Subfamily of HECT E3 Ubiquitin Ligases in Neurodevelopment and Neurodegeneration. Int. J. Mol. Sci. 2022, 23, 3882. [Google Scholar] [CrossRef]
- Li, Y.; Ozaki, T.; Kikuchi, H.; Yamamoto, H.; Ohira, M.; Nakagawara, A. A novel HECT-type E3 ubiquitin protein ligase NEDL1 enhances the p53-mediated apoptotic cell death in its catalytic activity-independent manner. Oncogene 2008, 27, 3700–3709. [Google Scholar] [CrossRef] [Green Version]
- Quiroga, M.; Rodríguez-Alonso, A.; Alfonsín, G.; Rodríguez, J.J.E.; Breijo, S.M.; Chantada, V.; Figueroa, A. Protein Degradation by E3 Ubiquitin Ligases in Cancer Stem Cells. Cancers 2022, 14, 990. [Google Scholar] [CrossRef]
- Favorov, A.V.; Andreewski, T.V.; Sudomoina, M.A.; Favorova, O.O.; Parmigiani, G.; Ochs, M.F. A Markov chain Monte Carlo technique for identification of combinations of allelic variants underlying complex diseases in humans. Genetics 2005, 171, 2113–2121. [Google Scholar] [CrossRef] [Green Version]
- Yazdani, U.; Terman, J.R. The semaphorins. Genome Biol. 2006, 7, 211. [Google Scholar] [CrossRef] [Green Version]
- de Wit, J.; Verhaagen, J. Role of semaphorins in the adult nervous system. Prog. Neurobiol. 2003, 71, 249–267. [Google Scholar] [CrossRef]
- Leighton, P.A.; Mitchell, K.J.; Goodrich, L.V.; Lu, X.; Pinson, K.; Scherz, P.; Skarnes, W.C.; Tessier-Lavigne, M. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature 2001, 410, 174–179. [Google Scholar] [CrossRef]
- Loria, R.; Bon, G.; Perotti, V.; Gallo, E.; Bersani, I.; Baldassari, P.; Porru, M.; Leonetti, C.; Di Carlo, S.; Visca, P.; et al. Sema6A and Mical1 control cell growth and survival of BRAFV600E human melanoma cells. Oncotarget 2015, 6, 2779–2793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, G.; Roshandel, D.; Sherva, R.; Monach, P.A.; Lu, E.Y.; Kung, T.; Carrington, K.; Zhang, S.S.; Pulit, S.L.; Ripke, S.; et al. Association of granulomatosis with polyangiitis (Wegener’s) with HLA–DPB1*04 and SEMA6A gene variants: Evidence from genome-wide analysis. Arthritis Rheum. 2013, 65, 2457–2468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Rahman, S.M.; Preuett, B.L. Genetic predictors of susceptibility to cutaneous fungal infections: A pilot genome wide association study to refine a candidate gene search. J. Dermatol. Sci. 2012, 67, 147–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, S.; Liu, Y.; Wu, H.; Liu, H.; Zeng, J.; Choi, M.Y.; Chen, H.; Gerhard, R.; Dong, M. Genome-wide CRISPR screen identifies semaphorin 6A and 6B as receptors for Paeniclostridium sordellii toxin TcsL. Cell Host Microbe. 2020, 27, 782–792.e7. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, Y.; Kang, S.; Kumanogoh, A. Axon guidance molecules in immunometabolic diseases. Inflamm. Regen. 2022, 42, 5. [Google Scholar] [CrossRef]
- Segarra, M.; Ohnuki, H.; Maric, D.; Salvucci, O.; Hou, X.; Kumar, A.; Li, X.; Tosato, G. Semaphorin 6A regulates angiogenesis by modulating VEGF signaling. Blood J. Am. Soc. Hematol. 2012, 120, 4104–4115. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.; Mu, X.; Yang, F.; Greenwald, E.; Park, J.W.; Jacob, E.; Zhang, C.Z.; Hur, S. Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation. Cell 2018, 172, 797–810.e13. [Google Scholar] [CrossRef] [Green Version]
- Lvovs, D.; Favorova, O.O.; Favorov, A.V. A polygenic approach to the study of polygenic diseases. Acta Nat. 2012, 4, 59–71. [Google Scholar] [CrossRef]
- Peze-Heidsieck, E.; Bonnifet, T.; Znaidi, R.; Ravel-Godreuil, C.; Massiani-Beaudoin, O.; Joshi, R.L.; Fuchs, J. Retrotransposons as a Source of DNA Damage in Neurodegeneration. Front. Aging Neurosci. 2022, 13, 786897. [Google Scholar] [CrossRef]
- Dyussenbayev, A. Age periods of human life. Adv. Soc. Sci. Res. J. 2017, 4, 258–263. [Google Scholar] [CrossRef]
| Genotype/ Allele | Young (18–44 Years Old) | Middle-Aged (45–59 Years Old) | Elderly (60–74 Years Old) | Old Seniors (75–89 Years Old) | Long-Livers (90–113 Years Old) | PHWEY |
|---|---|---|---|---|---|---|
| ACE Ya5ACE | ||||||
| DD | 28.95 | 27.31 | 29.11 | 29.52 | 25.34 | 3.04 × 10−1 |
| ID | 47.72 | 54.63 | 47.15 | 45.93 | 51.13 | |
| II | 23.32 | 18.06 | 23.73 | 24.55 | 23.53 | |
| D | 52.82 | 54.63 | 52.69 | 52.48 | 50.9 | |
| I | 47.18 | 45.37 | 47.31 | 47.52 | 49.1 | |
| HECW1 Ya5NBC182 | ||||||
| DD | 8.23 | 16.79 | 17.04 | 12.30 | 9.15 | 2.86 × 10−1 |
| ID | 46.75 | 39.69 | 42.59 | 41.73 | 46.41 | |
| II | 45.02 | 43.51 | 40.37 | 45.97 | 44.44 | |
| D | 31.60 | 36.64 | 38.33 | 33.17 | 32.35 | |
| I | 68.40 | 63.36 | 61.67 | 66.83 | 67.65 | |
| SEMA6A Yb8NBC597 | ||||||
| DD | 61.02 | 66.91 | 66.42 | 62.43 | 56.32 | 2.61 × 10−1 |
| ID | 35.83 | 28.06 | 28.73 | 31.98 | 33.91 | |
| II | 3.15 | 5.04 | 4.85 | 5.59 | 9.77 | |
| D | 78.94 | 80.94 | 80.78 | 78.42 | 73.28 | |
| I | 21.06 | 19.06 | 19.22 | 21.58 | 26.72 | |
| CDH4 Yb8NBC516 | ||||||
| DD | 17.80 | 25.25 | 18.98 | 14.44 | 26.22 | 6.60 × 10−2 |
| ID | 40.84 | 41.41 | 41.20 | 44.61 | 43.9 | |
| II | 41.36 | 33.33 | 39.81 | 40.95 | 29.88 | |
| D | 38.22 | 45.96 | 39.58 | 36.75 | 48.17 | |
| I | 61.78 | 54.04 | 60.42 | 63.25 | 51.83 | |
| STK38L Ya5ac2145 | ||||||
| DD | 80.72 | 82.55 | 81.65 | 79.96 | 76.97 | 1.71 × 10−1 |
| ID | 17.27 | 15.44 | 15.73 | 18.34 | 21.91 | |
| II | 2.01 | 2.01 | 2.62 | 1.70 | 1.12 | |
| D | 89.36 | 90.27 | 89.51 | 89.13 | 87.92 | |
| I | 10.64 | 9.73 | 10.49 | 10.87 | 12.08 | |
| PKHD1L1 Yb8AC702 | ||||||
| DD | 22.35 | 20.00 | 19.21 | 27.50 | 30.2 | 6.17 × 10−1 |
| ID | 51.76 | 58.13 | 60.93 | 52.17 | 49.5 | |
| II | 25.88 | 21.88 | 19.87 | 20.33 | 20.3 | |
| D | 48.24 | 49.06 | 49.67 | 53.58 | 54.95 | |
| I | 51.76 | 50.94 | 50.33 | 46.42 | 45.05 | |
| TEAD1 Ya5ac2013 | ||||||
| DD | 25.50 | 28.38 | 26.09 | 27.45 | 23.63 | 3.12 × 10−1 |
| ID | 46.61 | 42.57 | 47.83 | 44.72 | 45.6 | |
| II | 27.89 | 29.05 | 26.09 | 27.83 | 30.77 | |
| D | 48.80 | 49.66 | 50.00 | 49.81 | 46.43 | |
| I | 51.20 | 50.34 | 50.00 | 50.19 | 53.57 | |
| PLAT TPA25 | ||||||
| DD | 31.70 | 32.28 | 29.05 | 30.76 | 33.33 | 5.10 × 10−2 |
| ID | 44.12 | 50.26 | 43.24 | 46.71 | 40.38 | |
| II | 24.18 | 17.46 | 27.70 | 22.53 | 26.29 | |
| D | 53.76 | 57.41 | 50.68 | 54.11 | 53.52 | |
| I | 46.24 | 42.59 | 49.32 | 45.89 | 46.48 | |
| COL13A1 Ya5ac1986 | ||||||
| DD | 5.63 | 10.06 | 7.77 | 7.06 | 9.42 | 1.00 |
| ID | 36.62 | 34.32 | 42.07 | 37.27 | 32.74 | |
| II | 57.75 | 55.62 | 50.16 | 55.67 | 57.85 | |
| D | 23.94 | 27.22 | 28.80 | 25.69 | 25.78 | |
| I | 76.06 | 72.78 | 71.20 | 74.31 | 74.22 | |
| LAMA2 Ya5-MLS19 | ||||||
| DD | 29.59 | 36.51 | 33.23 | 37.21 | 24.45 | 7.25 × 10−1 |
| ID | 48.64 | 41.80 | 47.28 | 44.87 | 60.26 | |
| II | 21.77 | 21.69 | 19.49 | 17.92 | 15.28 | |
| D | 53.91 | 57.41 | 56.87 | 59.65 | 54.59 | |
| I | 46.09 | 42.59 | 43.13 | 40.35 | 45.41 | |
| Gene Alu Element | Genotype/ Allele | Reference Group | OR (CIOR) | P | Sex-Adjusted OR (CIOR) | Sex-Adjusted P |
|---|---|---|---|---|---|---|
| Long-livers | ||||||
| LAMA2 Ya5-MLS19 | DD | Middle-aged | 0.563 (0.369–0.859) | 8.00 × 10−3 | 0.568 (0.358–0.903) | 1.70 × 10−2 |
| Elderly | 0.651 (0.444–0.953) | 2.70 × 10−2 | 0.648 (0.440–0.956) | 2.90 × 10−2 | ||
| Old seniors | 0.546 (0.389–0.768) | 5.00 × 10−4 * | 0.554 (0.391–0.785) | 1.00 × 10−3 | ||
| ID | Young | 1.633 (1.145–2.328) | 7.00 × 10−3 | 1.491 (0.933–2.393) | 9.50 × 10−2 | |
| Middle-aged | 2.153 (1.449–3.201) | 1.49 × 10−4 * | 1.914 (1.237–2.959) | 4.00 × 10−3 | ||
| Elderly | 1.724 (1.214–2.448) | 2.00 × 10−3 | 1.726 (1.208–2.466) | 3.00 × 10−3 | ||
| Old seniors | 1.900 (1.391–2.596) | 5.51 × 10−4 * | 1.815 (1.318–2.501) | 2.66 × 10−4 | ||
| CDH4 Yb8NBC516 | II | Young | 0.604 (0.389–0.939) | 2.50 × 10−2 | 0.612 (0.345–1.083) | 9.2 × 10−2 |
| Old seniors | 0.614 (0.419–0.900) | 1.20 × 10−2 | 0.670 (0.453–0.989) | 4.40 × 10−2 | ||
| I | Young | 0.609 (0.366–1.013) | 5.60 × 10−2 | 0.680 (0.351–1.315) | 2.50 × 10−1 | |
| Elderly | 0.659 (0.405–1.072) | 9.30 × 10−2 | 0.659 (0.402–1.080) | 9.80 × 10−2 | ||
| Old seniors | 0.475 (0.308–0.733) | 1.00 × 10−3 * | 0.514 (0.330–0.800) | 3.00 × 10−3 | ||
| DD | Old seniors | 2.106 (1.365–3.249) | 1.00 × 10−3 | 1.946 (1.250–3.029) | 3.00 × 10−3 | |
| SEMA6A Yb8NBC597 | DD | Elderly | 0.652 (0.440–0.965) | 3.30 × 10−2 | 0.640 (0.430–0.955) | 2.90 × 10−2 |
| D | Elderly | 0.471 (0.223–0.996) | 4.90 × 10−2 | 0.487 (0.228–1.042) | 6.40 × 10−2 | |
| II | Young | 3.330 (1.404–7.899) | 6.00 × 10−3 | 3.794 (1.257–11.449) | 1.80 × 10−2 | |
| PKHD1L1 Yb8AC702 | DD | Middle-aged | 1.730 (1.060–2.825) | 2.80 × 10−2 | 2.022 (1.168–3.500) | 1.20 × 10−2 |
| Elderly | 1.820 (1.202–2.756) | 5.00 × 10−3 | 1.780 (1.166–2.718) | 8.00 × 10−3 | ||
| ID | Elderly | 0.629 (0.439–0.901) | 1.10 × 10−2 | 0.651 (0.451–0.940) | 2.20 × 10−2 | |
| PLAT TPA25 | II | Middle-aged | 1.686 (1.039–2.735) | 3.40 × 10−2 | 1.634 (0.953–2.803) | 7.40 × 10−2 |
| COL13A1 Ya5ac1986 | ID | Elderly | 0.670 (0.468–0.960) | 2.90 × 10−2 | 0.672 (0.466–0.968) | 3.30 × 10−2 |
| HECW1 Ya5NBC182 | DD | Elderly | 0.490 (0.260–0.925) | 2.80 × 10−2 | 0.473 (0.249–0.903) | 2.30 × 10−2 |
| Old seniors | ||||||
| ACE Ya5ACE | ID | Middle-aged | 0.706 (0.518–0.960) | 2.70 × 10−2 | 0.706 (0.518–0.962) | 2.70 × 10−2 |
| LAMA2 Ya5-MLS19 | DD | Young | 1.410 (1.048–1.897) | 2.30 × 10−2 | 1.368 (0.994–1.882) | 5.50 × 10−2 |
| CDH4 Yb8NBC516 | DD | Middle-aged | 0.500 (0.296–0.842) | 9.00 × 10−3 | 0.479 (0.283–0.812) | 6.00 × 10−3 |
| PKHD1L1 Yb8AC702 | DD | Elderly | 1.596 (1.138–2.237) | 7.00 × 10−3 | 1.591 (1.134–2.231) | 7.00 × 10−3 |
| ID | Elderly | 0.699 (0.528–0.927) | 1.30 × 10−2 | 0.708 (0.534–0.939) | 1.70 × 10−2 | |
| Elderly | ||||||
| HECW1 Ya5NBC182 | DD | Young | 2.291 (1.300–4.038) | 4.00 × 10−3 | 2.459 (1.306–4.631) | 5.00 × 10−3 |
| PKHD1L1 Yb8AC702 | ID | Young | 1.453 (1.037–2.036) | 3.00 × 10−2 | 1.380 (0.929–2.050) | 1.10 × 10−1 |
| PLAT TPA25 | II | Middle-aged | 1.811 (1.151–2.851) | 1.00 × 10−2 | 1.747 (1.095–2.787) | 1.90 × 10−2 |
| Combinations | Compared Age Periods # | P | PBonf | OR | CIOR | ||||
|---|---|---|---|---|---|---|---|---|---|
| 18–74 | 18–89 | 60–89 | 75–89 | 90–113 | |||||
| LAMA2 Ya5-MLS19*ID + CDH4 Yb8NBC516*DD + HECW1 Ya5NBC182*D | 3.77 | 15.22 | 1.70 × 10−6 | 9.00 × 10−3 | 4.58 | 2.56–8.21 | |||
| LAMA2 Ya5-MLS19*D + CDH4 Yb8NBC516*DD + HECW1 Ya5NBC182*D | 4.76 | 17.39 | 1.09 × 10−5 | 2.60 × 10−2 | 4.21 | 2.23–7.96 | |||
| LAMA2 Ya5-MLS19*D + CDH4 Yb8NBC516*DD + ACE Ya5ACE*I | 6.02 | 19.08 | 7.76 × 10−6 | 1.90 × 10−2 | 3.68 | 2.09–6.49 | |||
| LAMA2 Ya5-MLS19*ID + CDH4 Yb8NBC516*D + SEMA6A Yb8NBC597*I | 8.37 | 22.22 | 1.08 × 10−5 | 2.00 × 10−2 | 3.13 | 1.90–5.16 | |||
| LAMA2 Ya5-MLS19*ID + CDH4 Yb8NBC516*D | 28.12 | 46.58 | 3.76 × 10−6 | 1.90 × 10−2 | 2.23 | 1.59–3.13 | |||
| LAMA2 Ya5-MLS19*ID + HECW1 Ya5NBC182*I | 40.75 | 60.27 | 8.29 × 10−6 | 1.90 × 10−2 | 2.21 | 1.55–3.15 | |||
| Age Group | Age Range, Years Old | Sample Size, n | Mean Age ± SD, Years Old | Male/Female, n (%) |
|---|---|---|---|---|
| Young | 18–44 | 542 | 31.92 ± 7.82 | 390/152 (71.96/28.04) |
| Middle-aged | 45–59 | 261 | 50.87 ± 4.42 | 152/109 (58.04/41.76) |
| Elderly | 60–74 | 321 | 68.39 ± 3.93 | 107/214 (33.33/66.67) |
| Old seniors | 75–89 | 693 | 80.48 ± 3.74 | 287/406 (41.41/58.59) |
| Long-livers | 90–113 | 237 | 93.16 ± 2.97 | 39/198 (16.46/83.54) |
| Total | 18–113 | 2054 | 63.48 ± 22.65 | 975/1079 (47.47/52.53) |
| Alu Element | Gene, Chromosome Location | Primers | Annealing Temperature (°C) | Alleles (Fragment Length, bp) |
|---|---|---|---|---|
| Ya5ACE | ACE 17q23.3 | F 5’-ctg gag acc act ccc atc ctt tct-3’ R 5’-gat gtg gcc atc aca ttc gtc aga t-3’ | 68 | I (490) D (190) |
| Ya5NBC182 | HECW1 7p13 | F 5′-gaa gga cta tgt agt tgc aga agc-3′ R 5′-aac cca gtg gaa aca gaa gat g-3′ | 64 | I (563) D (287) |
| Yb8NBC597 | SEMA6A 5q23.1 | F 5′-tga ggt gtt gca gac gat gt-3′ R 5′-cgc atg ctt tag aga ata ccc-3′ | 63 | I (429) D (108) |
| Yb8NBC516 | CDH4 20q13.33 | F 5′-ggg ctc agg gat act atg ctc-3′ R 5′-gcc tag gcc tac cac tca ga-3′ | 60 | I (445) D (124) |
| Ya5ac2145 | STK38L 12p11.23 | F 5′-tgt tct aat gac cat gcc tac tt-3′ R 5′-tgc ctt tag gaa gct aca gat tta-3′ | 60 | I (465) D (135) |
| Yb8AC702 | PKHD1L1 8q23.2 | F 5′-tgt ttg gaa ata agc caa aca at-3′ R 5′-ggg tag caa cct ttt tca tct tt-3′ | 60 | I (482) D (161) |
| Ya5ac2013 | TEAD1 11p15.2 | F 5′-tgg cag att ctg act ggc ta-3′ R 5′-cac gta agg tga aaa ggg ga-3′ | 60 | I (489) D (212) |
| TPA25 | PLAT 8p11.21 | F 5′-caa cca atg aaa acc act ga-3′ R 5′-gtt ctc ctg aca tct tta ttg-3′ | 60 | I (518) D (217) |
| Ya5ac1986 | COL13A1 10q22.1 | F 5′-tct agt ggg atg agg ata ac-3′ R 5′-tgt gcc atg ggg taa gaa ac-3′ | 60 | I (431) D (134) |
| Ya5-MLS19 | LAMA2 6q22.33 | F 5′-cta tga cgg agt aaa aag aag t-3′ R 5′-gaa aga gtg cca acc ctg tcc-3′ | 63 (7 cycles) 60 (22 cycles) | I (401) D (106) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erdman, V.V.; Karimov, D.D.; Tuktarova, I.A.; Timasheva, Y.R.; Nasibullin, T.R.; Korytina, G.F. Alu Deletions in LAMA2 and CDH4 Genes Are Key Components of Polygenic Predictors of Longevity. Int. J. Mol. Sci. 2022, 23, 13492. https://doi.org/10.3390/ijms232113492
Erdman VV, Karimov DD, Tuktarova IA, Timasheva YR, Nasibullin TR, Korytina GF. Alu Deletions in LAMA2 and CDH4 Genes Are Key Components of Polygenic Predictors of Longevity. International Journal of Molecular Sciences. 2022; 23(21):13492. https://doi.org/10.3390/ijms232113492
Chicago/Turabian StyleErdman, Vera V., Denis D. Karimov, Ilsia A. Tuktarova, Yanina R. Timasheva, Timur R. Nasibullin, and Gulnaz F. Korytina. 2022. "Alu Deletions in LAMA2 and CDH4 Genes Are Key Components of Polygenic Predictors of Longevity" International Journal of Molecular Sciences 23, no. 21: 13492. https://doi.org/10.3390/ijms232113492
APA StyleErdman, V. V., Karimov, D. D., Tuktarova, I. A., Timasheva, Y. R., Nasibullin, T. R., & Korytina, G. F. (2022). Alu Deletions in LAMA2 and CDH4 Genes Are Key Components of Polygenic Predictors of Longevity. International Journal of Molecular Sciences, 23(21), 13492. https://doi.org/10.3390/ijms232113492