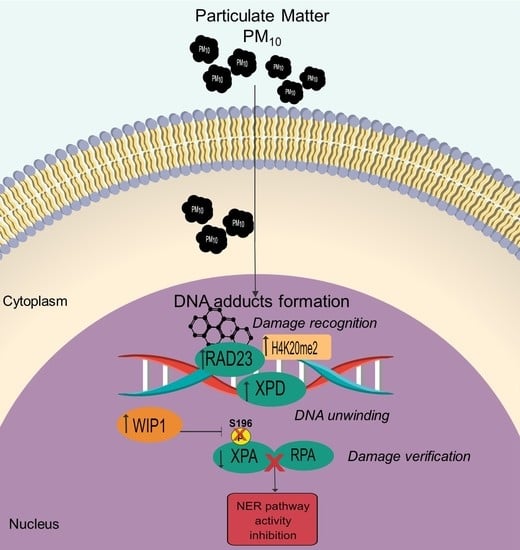
Nucleotide Excision Repair Pathway Activity Is Inhibited by Airborne Particulate Matter (PM10) through XPA Deregulation in Lung Epithelial Cells
Abstract
:1. Introduction
2. Results
2.1. PM10 Induced the Formation of BPDE–DNA Adducts
2.2. PM10 Deregulated the RAD23, XPD, and XPA Proteins Used in the Recognition and Verification Step of the NER Pathway
2.3. PM10 Induced Nuclear Recruitment (H4K20me2) and Dephosphorylation of XPA Associated with WIP1 Increase
2.4. PM10 Impaired the Formation of the XPA-RPA Complex
2.5. The NER Pathway Was Inactive in Cells Exposed to PM10
3. Discussion
4. Materials and Methods
4.1. PM10 Collection
4.2. Cell Culture and PM10 Exposure
4.3. Measurement of the Benzo(a)pyrene-7,8-diol-9,10-epoxide-DNA Adducts (BPDE-DNA Adduct)
4.4. Evaluation of the Total Protein Levels of the NER Pathway
4.5. Measurements of Nuclear Protein Levels of the NER Pathway
4.6. Detection of the XPA-RPA Protein Complex
4.7. Measurement of NER Activity
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bai, L.; Shin, S.; Burnett, R.T.; Kwong, J.C.; Hystad, P.; van Donkelaar, A.; Goldberg, M.S.; Lavigne, E.; Weichenthal, S.; Martin, R.V.; et al. Exposure to Ambient Air Pollution and the Incidence of Lung Cancer and Breast Cancer in the Ontario Population Health and Environment Cohort. Int. J. Cancer 2020, 146, 2450–2459. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Guo, S.; Xu, R.; Ye, T.; Li, S.; Sim, M.R.; Abramson, M.J.; Guo, Y. Cohort Studies of Long-Term Exposure to Outdoor Particulate Matter and Risks of Cancer: A Systematic Review and Meta-Analysis. Innovation 2021, 2, 100035. [Google Scholar] [CrossRef] [PubMed]
- Loomis, D.; Huang, W.; Chen, G. The International Agency for Research on Cancer (IARC) Evaluation of the Carcinogenicity of Outdoor Air Pollution: Focus on China. Chin. J. Cancer 2014, 33, 189–196. [Google Scholar] [CrossRef]
- Turner, M.C.; Andersen, Z.J.; Baccarelli, A.; Diver, W.R.; Gapstur, S.M.; Pope, C.A.; Prada, D.; Samet, J.; Thurston, G.; Cohen, A. Outdoor Air Pollution and Cancer: An Overview of the Current Evidence and Public Health Recommendations. CA Cancer J. Clin. 2020, 70, 460–479. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, L.-W.; Huang, J.-J.; Song, F.-J.; Zhang, L.-P.; Qian, Z.-M.; Trevathan, E.; Mao, H.-J.; Han, B.; Vaughn, M.; et al. Long-Term Exposure to Urban Air Pollution and Lung Cancer Mortality: A 12-Year Cohort Study in Northern China. Sci. Total Environ. 2016, 571, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Pope, C.A., 3rd; Burnett, R.T.; Thun, M.J.; Calle, E.E.; Krewski, D.; Ito, K.; Thurston, G.D. Lung Cancer, Cardiopulmonary Mortality, and Long-Term Exposure to Fine Particulate Air Pollution. JAMA 2002, 287, 1132–1141. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Li, L.; Hu, L. Correlation Analysis of PM10 and the Incidence of Lung Cancer in Nanchang, China. Int. J. Environ. Res. Public Health 2017, 14, 1253. [Google Scholar] [CrossRef] [Green Version]
- Moon, D.H.; Kwon, S.O.; Kim, S.-Y.; Kim, W.J. Air Pollution and Incidence of Lung Cancer by Histological Type in Korean Adults: A Korean National Health Insurance Service Health Examinee Cohort Study. Int. J. Environ. Res. Public Health 2020, 17, 915. [Google Scholar] [CrossRef] [Green Version]
- Consonni, D.; Carugno, M.; De Matteis, S.; Nordio, F.; Randi, G.; Bazzano, M.; Caporaso, N.E.; Tucker, M.A.; Bertazzi, P.A.; Pesatori, A.C.; et al. Outdoor Particulate Matter (PM10) Exposure and Lung Cancer Risk in the EAGLE Study. PLoS ONE 2018, 13, e0203539. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-H.; Wu, C.-D.; Chiang, H.-C.; Chu, D.; Lee, K.-Y.; Lin, W.-Y.; Yeh, J.-I.; Tsai, K.-W.; Guo, Y.L. The Effects of Fine and Coarse Particulate Matter on Lung Function among the Elderly. Sci. Rep. 2019, 9, 14790. [Google Scholar] [CrossRef]
- Yoda, Y.; Takagi, H.; Wakamatsu, J.; Ito, T.; Nakatsubo, R.; Horie, Y.; Hiraki, T.; Shima, M. Stronger Association between Particulate Air Pollution and Pulmonary Function among Healthy Students in Fall than in Spring. Sci. Total Environ. 2019, 675, 483–489. [Google Scholar] [CrossRef]
- Chirino, Y.I.; Sánchez-Pérez, Y.; Osornio-Vargas, Á.R.; Morales-Bárcenas, R.; Gutiérrez-Ruíz, M.C.; Segura-García, Y.; Rosas, I.; Pedraza-Chaverri, J.; García-Cuellar, C.M. PM10 Impairs the Antioxidant Defense System and Exacerbates Oxidative Stress Driven Cell Death. Toxicol. Lett. 2010, 193, 209–216. [Google Scholar] [CrossRef]
- Valavanidis, A.; Vlachogianni, T.; Fiotakis, K. Comparative Study of the Formation of Oxidative Damage Marker 8-Hydroxy-2′-Deoxyguanosine (8-OHdG) Adduct from the Nucleoside 2′-Deoxyguanosine by Transition Metals and Suspensions of Particulate Matter in Relation to Metal Content and Redox Reactivity. Free Radic. Res. 2005, 39, 1071–1081. [Google Scholar] [CrossRef]
- Krokan, H.E.; Bjørås, M. Base Excision Repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef]
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative Stress and Antioxidant Defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Błaszczyk, E.; Rogula-Kozłowska, W.; Klejnowski, K.; Fulara, I.; Mielżyńska-Švach, D. Polycyclic Aromatic Hydrocarbons Bound to Outdoor and Indoor Airborne Particles (PM2.5) and Their Mutagenicity and Carcinogenicity in Silesian Kindergartens, Poland. Air Qual. Atmos. Health 2016, 10, 389–400. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Wang, Z.; Qian, L.; Zhao, Z.; Zhang, C.; Fu, Y.; Li, J.; Zhang, C.; Lu, B.; Qian, J. Biological and Chemical Compositions of Atmospheric Particulate Matter during Hazardous Haze Days in Beijing. Environ. Sci. Pollut. Res. 2018, 25, 34540–34549. [Google Scholar] [CrossRef] [Green Version]
- Hadei, M.; Aboosaedi, Z.; Naddafi, K. Carcinogenic Risks and Chemical Composition of Particulate Matter Recovered by Two Methods: Wet and Dry Extraction. Environ. Monit. Assess. 2020, 192, 213–217. [Google Scholar] [CrossRef]
- Salcido-Neyoy, M.E.; Sánchez-Pérez, Y.; Osornio-Vargas, A.R.; Gonsebatt, M.E.; Meléndez-Zajgla, J.; Morales-Bárcenas, R.; Petrosyan, P.; Molina-Servin, E.D.; Vega, E.; Manzano-León, N.; et al. Induction of c-Jun by Air Particulate Matter (PM10) of Mexico City: Participation of Polycyclic Aromatic Hydrocarbons. Environ. Pollut. 2015, 203, 175–182. [Google Scholar] [CrossRef]
- Lepers, C.; André, V.; Dergham, M.; Billet, S.; Verdin, A.; Garcon, G.; Dewaele, D.; Cazier, F.; Sichel, F.; Shirali, P. Xenobiotic Metabolism Induction and Bulky DNA Adducts Generated by Particulate Matter Pollution in BEAS-2B Cell Line: Geographical and Seasonal Influence. J. Appl. Toxicol. 2013, 34, 703–713. [Google Scholar] [CrossRef]
- Stiborová, M.; Moserová, M.; Černá, V.; Indra, R.; Dračínský, M.; Šulc, M.; Henderson, C.J.; Wolf, C.R.; Schmeiser, H.H.; Phillips, D.H.; et al. Cytochrome b5 and Epoxide Hydrolase Contribute to Benzo[a]pyrene-DNA Adduct Formation Catalyzed by CytoChrome P450 1A1 under Low NADPH: P450 Oxidoreductase Conditions. Toxicology 2014, 318, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Braithwaite, E.; Wu, X.; Wang, Z. Repair of DNA Lesions Induced by Polycyclic Aromatic Hydrocarbons in Human Cell-Free Extracts: Involvement of Two Excision Repair Mechanisms in Vitro. Carcinogenesis 1998, 19, 1239–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butkiewicz, D.; Rusin, M.; Pawlas, M.; Czarny, M.; Chorazy, M. Repair of DNA Damage Using Nucleotide Excision Repair (NER)-Relationship with Cancer Risk. Postepy Hig. I Med. Dosw. 2002, 56, 485–498. [Google Scholar]
- Hoeijmakers, J.H. Nucleotide Excision Repair II: From Yeast to Mammals. Trends Genet. 1993, 9, 211–217. [Google Scholar] [CrossRef] [Green Version]
- Chitale, S.; Richly, H. DICER- and MMSET-Catalyzed H4K20me2 Recruits the Nucleotide Excision Repair Factor XPA to DNA Damage Sites. J. Cell Biol. 2017, 217, 527–540. [Google Scholar] [CrossRef]
- Sugasawa, K.; Ng, J.M.; Masutani, C.; Iwai, S.; van der Spek, P.J.; Eker, A.P.; Hanaoka, F.; Bootsma, D.; Hoeijmakers, J.H. Xeroderma Pigmentosum Group C Protein Complex Is the Initiator of Global Genome Nucleotide Excision Repair. Mol. Cell 1998, 2, 223–232. [Google Scholar] [CrossRef]
- Kuper, J.; Braun, C.; Elias, A.; Michels, G.; Sauer, F.; Schmitt, D.R.; Poterszman, A.; Egly, J.-M.; Kisker, C. In TFIIH, XPD Helicase Is Exclusively Devoted to DNA Repair. PLoS Biol. 2014, 12, e1001954. [Google Scholar] [CrossRef] [Green Version]
- Volker, M.; Moné, M.J.; Karmakar, P.; van Hoffen, A.; Schul, W.; Vermeulen, W.; Hoeijmakers, J.H.; van Driel, R.; van Zeeland, A.A.; Mullenders, L.H. Sequential Assembly of the Nucleotide Excision Repair Factors In Vivo. Mol. Cell 2001, 8, 213–224. [Google Scholar] [CrossRef]
- Rechkunova, N.I.; Maltseva, E.A.; Lavrik, O.I. Post-Translational Modifications of Nucleotide Excision Repair Proteins and Their Role in the DNA Repair. Biochemistry 2019, 84, 1008–1020. [Google Scholar] [CrossRef]
- Lee, T.-H.; Park, J.-M.; Leem, S.-H.; Kang, T.-H. Coordinated Regulation of XPA Stability by ATR and HERC2 during Nucleotide Excision Repair. Oncogene 2012, 33, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.-A.; Slattery, S.D.; Moon, S.-H.; Darlington, Y.F.; Lu, X.; Donehower, L.A. The Oncogenic Phosphatase WIP1 Negatively Regulates Nucleotide Excision Repair. DNA Repair 2010, 9, 813–823. [Google Scholar] [CrossRef] [Green Version]
- Park, J.-M.; Kang, T.-H. Transcriptional and Posttranslational Regulation of Nucleotide Excision Repair: The Guardian of the Genome against Ultraviolet Radiation. Int. J. Mol. Sci. 2016, 17, 1840. [Google Scholar] [CrossRef] [PubMed]
- Shell, S.M.; Li, Z.; Shkriabai, N.; Kvaratskhelia, M.; Brosey, C.; Serrano, M.; Chazin, W.J.; Musich, P.; Zou, Y. Checkpoint Kinase ATR Promotes Nucleotide Excision Repair of UV-induced DNA Damage via Physical Interaction with Xeroderma Pigmentosum Group A. J. Biol. Chem. 2009, 284, 24213–24222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Lu, X.; Peterson, C.A.; Legerski, R.J. An Interaction between the DNA Repair Factor XPA and Replication Protein A Appears Essential for Nucleotide Excision Repair. Mol. Cell. Biol. 1995, 15, 5396–5402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borszéková Pulzová, L.; Ward, T.A.; Chovanec, M. XPA: DNA Repair Protein of Significant Clinical Importance. Int. J. Mol. Sci. 2020, 21, 2182. [Google Scholar] [CrossRef] [Green Version]
- Bai, H.; Wu, M.; Zhang, H.; Tang, G. Chronic Polycyclic Aromatic Hydrocarbon Exposure Causes DNA Damage and Genomic Instability in Lung Epithelial Cells. Oncotarget 2017, 8, 79034–79045. [Google Scholar] [CrossRef] [Green Version]
- Veglia, F.; Matullo, G.; Vineis, P. Bulky DNA Adducts and Risk of Cancer: A Meta-Analysis. Cancer Epidemiol. Biomark. Prev. 2003, 12, 157–160. [Google Scholar]
- Otteneder, M.; Lutz, W.K. Correlation of DNA Adduct Levels with Tumor Incidence: Carcinogenic Potency of DNA Adducts. Mutat. Res. Mol. Mech. Mutagen. 1999, 424, 237–247. [Google Scholar] [CrossRef]
- Cheng, L.; Spitz, M.R.; Hong, W.K.; Wei, Q. Reduced Expression Levels of Nucleotide Excision Repair Genes in Lung Cancer: A Case-Control Analysis. Carcinogenesis 2000, 21, 1527–1530. [Google Scholar] [CrossRef]
- Ide, F.; Iida, N.; Nakatsuru, Y.; Oda, H.; Tanaka, K.; Ishikawa, T. Mice Deficient in the Nucleotide Excision Repair Gene XPA Have Elevated Sensitivity to Benzo[a]pyrene Induction of Lung Tumors. Carcinogenesis 2000, 21, 1263–1265. [Google Scholar]
- Barnes, J.L.; Zubair, M.; John, K.; Poirier, M.C.; Martin, F.L. Carcinogens and DNA Damage. Biochem. Soc. Trans. 2018, 46, 1213–1224. [Google Scholar] [CrossRef] [Green Version]
- Topinka, J.; Schwarz, L.; Wiebel, F.; Černá, M.; Wolff, T. Genotoxicity of Urban Air Pollutants in the Czech Republic: Part II. DNA Adduct Formation in Mammalian Cells by Extractable Organic Matter. Mutat. Res. Toxicol. Environ. Mutagen. 2000, 469, 83–93. [Google Scholar] [CrossRef]
- Palli, D.; Saieva, C.; Munnia, A.; Peluso, M.; Grechi, D.; Zanna, I.; Caini, S.; Decarli, A.; Sera, F.; Masala, G. DNA Adducts and PM10 Exposure in Traffic-Exposed Workers and Urban Residents from the EPIC-Florence City Study. Sci. Total Environ. 2008, 403, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Hao, M.; Phalen, R.F.; Hinds, W.C.; Nel, A.E. Particulate Air Pollutants and Asthma: A Paradigm for the Role of Oxidative Stress in PM-Induced Adverse Health Effects. Clin. Immunol. 2003, 109, 250–265. [Google Scholar] [CrossRef] [PubMed]
- Ferecatu, I.; Borot, M.-C.; Bossard, C.; Leroux, M.; Boggetto, N.; Marano, F.; Baeza-Squiban, A.; Andreau, K. Polycyclic Aromatic Hydrocarbon Components Contribute to the Mitochondria-Antiapoptotic Effect of Fine Particulate Matter on Human Bronchial Epithelial Cells via the Aryl Hydrocarbon Receptor. Part. Fibre Toxicol. 2010, 7, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piberger, A.L.; Krüger, C.T.; Strauch, B.M.; Schneider, B.; Hartwig, A. BPDE-Induced Genotoxicity: Relationship between DNA Adducts, Mutagenicity in the in Vitro PIG-A Assay, and the Transcriptional Response to DNA Damage in TK6 Cells. Arch. Toxicol. 2017, 92, 541–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosskopf, C.; Schwerdtle, T.; Mullenders, L.H.F.; Hartwig, A. Antimony Impairs Nucleotide Excision Repair: XPA and XPE as Potential Molecular Targets. Chem. Res. Toxicol. 2010, 23, 1175–1183. [Google Scholar] [CrossRef]
- Schwerdtle, T.; Ebert, F.; Thuy, C.; Richter, C.; Mullenders, L.H.F.; Hartwig, A. Genotoxicity of Soluble and Particulate Cadmium Compounds: Impact on Oxidative DNA Damage and Nucleotide Excision Repair. Chem. Res. Toxicol. 2010, 23, 432–442. [Google Scholar] [CrossRef]
- Shi, Q.; Maas, L.; Veith, C.; Van Schooten, F.J.; Godschalk, R.W. Acidic Cellular Microenvironment Modifies Carcinogen-Induced DNA Damage and Repair. Arch. Toxicol. 2016, 91, 2425–2441. [Google Scholar] [CrossRef] [Green Version]
- Riedl, T.; Hanaoka, F.; Egly, J. The Comings and Goings of Nucleotide Excision Repair Factors on Damaged DNA. EMBO J. 2003, 22, 5293–5303. [Google Scholar] [CrossRef] [Green Version]
- Krasikova, Y.S.; Rechkunova, N.I.; Maltseva, E.A.; Lavrik, O.I. RPA and XPA Interaction with DNA Structures Mimicking Intermediates of the Late Stages in Nucleotide Excision Repair. PLoS ONE 2018, 13, e0190782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Hu, Z.; Zhang, Y.; Gou, X.; Mu, Y.; Wang, L.; Xie, X.-Q. Metal Binding Mediated Conformational Change of XPA Protein: A Potential Cytotoxic Mechanism of Nickel in the Nucleotide Excision Repair. J. Mol. Model. 2016, 22, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartwig, A.; Asmuss, M.; Ehleben, I.; Herzer, U.; Kostelac, D.; Pelzer, A.; Schwerdtle, T.; Bürkle, A. Interference by Toxic Metal Ions with DNA Repair Processes and Cell Cycle Control: Molecular Mechanisms. Environ. Health Perspect. 2002, 110 (Suppl. 5), 797–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopera, E.; Schwerdtle, T.; Hartwig, A.; Bal, W. Co (II) and Cd (II) Substitute for Zn (II) in the Zinc Finger Derived from the DNA Repair Protein XPA, Demonstrating a Variety of Potential Mechanisms of Toxicity. Chem. Res. Toxicol. 2004, 17, 1452–1458. [Google Scholar] [CrossRef] [PubMed]
- Jan, R.; Roy, R.; Bhor, R.; Pai, K.; Satsangi, P.G. Toxicological Screening of Airborne Particulate Matter in Atmosphere of Pune: Reactive Oxygen Species and Cellular Toxicity. Environ. Pollut. 2019, 261, 113724. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shell, S.M.; Yang, Z.; Zou, Y. Phosphorylation of Nucleotide Excision Repair Factor Xeroderma Pigmentosum Group A by Ataxia Telangiectasia Mutated and Rad3-Related–Dependent Checkpoint Pathway Promotes Cell Survival in Response to UV Irradiation. Cancer Res. 2006, 66, 2997–3005. [Google Scholar] [CrossRef] [Green Version]
- Fiscella, M.; Zhang, H.; Fan, S.; Sakaguchi, K.; Shen, S.; Mercer, W.E.; Woude, G.F.V.; O’Connor, P.M.; Appella, E. Wip1, a Novel Human Protein Phosphatase That Is Induced in Response to Ionizing radiation in a p53-Dependent Manner. Proc. Natl. Acad. Sci. USA 1997, 94, 6048–6053. [Google Scholar] [CrossRef] [Green Version]
- Bai, F.; Zhou, H.; Fu, Z.; Xie, J.; Hu, Y.; Nie, S. NF-κB-Induced WIP1 Expression Promotes Colorectal Cancer Cell Proliferation through mTOR Signaling. Biomed. Pharmacother. 2018, 99, 402–410. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, H.; Zhu, G.; Liang, J.; Chen, N.; Yang, Y.; Liang, X.; Cai, H.; Liu, W. Association between Overexpression of Wip1 and Prognosis of Patients with Non-Small Cell Lung Cancer. Oncol. Lett. 2016, 11, 2365–2370. [Google Scholar] [CrossRef] [Green Version]
- Tanoue, K.; Jenkins, L.M.M.; Durell, S.R.; Debnath, S.; Sakai, H.; Tagad, H.D.; Ishida, K.; Appella, E.; Mazur, S.J. Binding of a Third Metal Ion by the Human Phosphatases PP2Cα and Wip1 Is Required for Phosphatase Activity. Biochemistry 2013, 52, 5830–5843. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y. Serine/Threonine Phosphatases: Mechanism through Structure. Cell 2009, 139, 468–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.-G.; Liu, Y.; Mao, L.Y.; Zhang, J.-T.; Zou, Y. Dimerization of Human XPA and Formation of XPA2−RPA Protein Complex. Biochemistry 2002, 41, 13012–13020. [Google Scholar] [CrossRef] [PubMed]
- Topolska-Woś, A.M.; Sugitani, N.; Cordoba, J.J.; Le Meur, K.V.; Le Meur, R.; Kim, H.S.; Yeo, J.-E.; Rosenberg, D.; Hammel, M.; Schärer, O.D.; et al. A Key Interaction with RPA Orients XPA in NER Complexes. Nucleic Acids Res. 2020, 48, 2173–2188. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Pérez, Y.; Chirino, Y.I.; Vargas, A.O.; Morales-Bárcenas, R.; Gutierrez-Ruiz, M.C.; Vázquez-López, I.; Garcia-Cuellar, C.M. DNA Damage Response of A549 Cells Treated with Particulate Matter (PM10) of Urban Air Pollutants. Cancer Lett. 2009, 278, 192–200. [Google Scholar] [CrossRef]
- Calderón-Garcidueñas, L.; Herrera-Soto, A.; Jury, N.; Maher, B.A.; González-Maciel, A.; Reynoso-Robles, R.; Ruiz-Rudolph, P.; van Zundert, B.; Varela-Nallar, L. Reduced Repressive Epigenetic Marks, Increased DNA Damage and Alzheimer’s Disease Hallmarks in the Brain of Humans and Mice Exposed to Particulate Urban Air Pollution. Environ. Res. 2020, 183, 109226. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez-Andrade, M.; Sánchez-Pérez, Y.; Chirino, Y.I.; Morales-Bárcenas, R.; García-Cuellar, C.M. Long Non-Coding RNA NORAD Upregulation Induced by Airborne Particulate Matter (PM10) Exposure Leads to Aneuploidy in A549 Lung Cells. Chemosphere 2020, 266, 128994. [Google Scholar] [CrossRef]
- Quezada-Maldonado, E.M.; Sánchez-Pérez, Y.; Chirino, Y.I.; García-Cuellar, C.M. Airborne Particulate Matter Induces Oxidative Damage, DNA Adduct Formation and Alterations in DNA Repair Pathways. Environ. Pollut. 2021, 287, 117313. [Google Scholar] [CrossRef]
- Papadopoulos, N.; Lindblom, A. Molecular Basis of HNPCC: Mutations of MMR Genes. Hum. Mutat. 1997, 10, 89–99. [Google Scholar] [CrossRef]
- Yoshioka, K.-I.; Kusumoto-Matsuo, R.; Matsuno, Y.; Ishiai, M. Genomic Instability and Cancer Risk Associated with Erroneous DNA Repair. Int. J. Mol. Sci. 2021, 22, 12254. [Google Scholar] [CrossRef]
- Langie, S.A.S.; Koppen, G.; Desaulniers, D.; Al-Mulla, F.; Altemaimi, R.; Amedei, A.; Azqueta, A.; Bisson, W.H.; Brown, D.; Brunborg, G.; et al. Causes of Genome Instability: The Effect of Low Dose Chemical Exposures in Modern Society. Carcinogenesis 2015, 36, S61–S88. [Google Scholar] [CrossRef]
- Wilhelm, T.; Said, M.; Naim, V. DNA Replication Stress and Chromosomal Instability: Dangerous Liaisons. Genes 2020, 11, 642. [Google Scholar] [CrossRef] [PubMed]
- Chirino, Y.I.; García-Cuellar, C.M.; García-García, C.; Soto-Reyes, E.; Osornio-Vargas, Á.R.; Herrera, L.A.; López-Saavedra, A.; Miranda, J.; Quintana-Belmares, R.; Pérez, I.R.; et al. Airborne Particulate Matter in Vitro Exposure Induces Cytoskeleton Remodeling through Activation of the ROCK-MYPT1-MLC Pathway in A549 Epithelial Lung Cells. Toxicol. Lett. 2017, 272, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Quezada-Maldonado, E.M.; Sánchez-Pérez, Y.; Chirino, Y.I.; Vaca-Paniagua, F.; García-Cuellar, C.M. miRNAs Deregulation in Lung Cells Exposed to Airborne Particulate Matter (PM10) Is Associated with Pathways Deregulated in Lung Tumors. Environ. Pollut. 2018, 241, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Abbas, I.; Garcon, G.; Saint-Georges, F.; Andre, V.; Gosset, P.; Billet, S.; Le Goff, J.; Verdin, A.; Mulliez, P.; Sichel, F.; et al. Polycyclic Aromatic Hydrocarbons within Airborne Particulate Matter (PM2.5) Produced DNA Bulky Stable Adducts in a Human Lung Cell Coculture Model. J. Appl. Toxicol. 2011, 33, 109–119. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.R.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Harbor, C.S., Ed.; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 1989. [Google Scholar]
- Morales-Bárcenas, R.; Chirino, Y.I.; Sánchez-Pérez, Y.; Osornio-Vargas, Á.R.; Melendez-Zajgla, J.; Rosas, I.; García-Cuellar, C.M. Particulate Matter (PM10) Induces Metalloprotease Activity and Invasion in Airway Epithelial Cells. Toxicol. Lett. 2015, 237, 167–173. [Google Scholar] [CrossRef]
- Wienholz, F.; Vermeulen, W.; Marteijn, J.A. Amplification of Unscheduled DNA Synthesis Signal Enables Fluorescence-Based Single Cell Quantification of Transcription-Coupled Nucleotide Excision Repair. Nucleic Acids Res. 2017, 45, e68. [Google Scholar] [CrossRef] [Green Version]
- Peter, F. Guengerich Mechanisms of Drug Toxicity and Relevance to Pharmaceutical Development. Drug Metab. Pharmacokinet. 2011, 26, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Salic, A.; Mitchison, T.J. A Chemical Method for Fast and Sensitive Detection of DNA Synthesis in Vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 2415–2420. [Google Scholar] [CrossRef] [Green Version]
- Bendjennat, M.; Boulaire, J.; Jascur, T.; Brickner, H.; Barbier, V.; Sarasin, A.; Fotedar, A.; Fotedar, R. UV Irradiation Triggers Ubiquitin-Dependent Degradation of p21WAF1 to Promote DNA Repair. Cell 2003, 114, 599–610. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quezada-Maldonado, E.M.; Chirino, Y.I.; Gonsebatt, M.E.; Morales-Bárcenas, R.; Sánchez-Pérez, Y.; García-Cuellar, C.M. Nucleotide Excision Repair Pathway Activity Is Inhibited by Airborne Particulate Matter (PM10) through XPA Deregulation in Lung Epithelial Cells. Int. J. Mol. Sci. 2022, 23, 2224. https://doi.org/10.3390/ijms23042224
Quezada-Maldonado EM, Chirino YI, Gonsebatt ME, Morales-Bárcenas R, Sánchez-Pérez Y, García-Cuellar CM. Nucleotide Excision Repair Pathway Activity Is Inhibited by Airborne Particulate Matter (PM10) through XPA Deregulation in Lung Epithelial Cells. International Journal of Molecular Sciences. 2022; 23(4):2224. https://doi.org/10.3390/ijms23042224
Chicago/Turabian StyleQuezada-Maldonado, Ericka Marel, Yolanda I. Chirino, María Eugenia Gonsebatt, Rocío Morales-Bárcenas, Yesennia Sánchez-Pérez, and Claudia M. García-Cuellar. 2022. "Nucleotide Excision Repair Pathway Activity Is Inhibited by Airborne Particulate Matter (PM10) through XPA Deregulation in Lung Epithelial Cells" International Journal of Molecular Sciences 23, no. 4: 2224. https://doi.org/10.3390/ijms23042224
APA StyleQuezada-Maldonado, E. M., Chirino, Y. I., Gonsebatt, M. E., Morales-Bárcenas, R., Sánchez-Pérez, Y., & García-Cuellar, C. M. (2022). Nucleotide Excision Repair Pathway Activity Is Inhibited by Airborne Particulate Matter (PM10) through XPA Deregulation in Lung Epithelial Cells. International Journal of Molecular Sciences, 23(4), 2224. https://doi.org/10.3390/ijms23042224