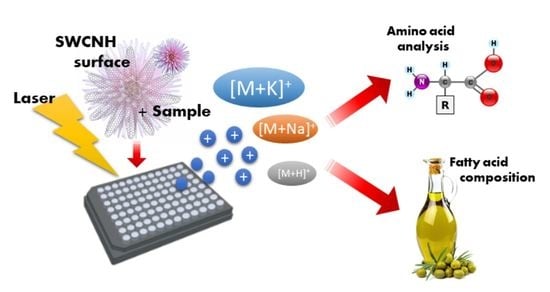
Single-Walled Carbon Nanohorns as Boosting Surface for the Analysis of Low-Molecular-Weight Compounds by SALDI-MS
Abstract
:1. Introduction
2. Results and Discussion
2.1. Evaluation of Characteristics and Performances of the MS-Surface
2.2. Analysis of Amino Acids
2.3. Analysis of Real Oil Samples
2.4. Future Perspectives
3. Material and Methods
3.1. Chemicals
3.2. Surface Preparation and Sample Deposition
3.3. MALDI Analysis
3.4. ICP-MS Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, C.-Y.; Lee, K.-C.; Kuo, Y.-L.; Chen, Y.-C. Revisiting the Quantitative Features of Surface-Assisted Laser Desorption/Ionization Mass Spectrometric Analysis. Philos. Trans. R. Soc. A 2016, 374, 20150379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, C.-K.; Chen, W.-T.; Chang, H.-T. Nanoparticle-Based Mass Spectrometry for the Analysis of Biomolecules. Chem. Soc. Rev. 2011, 40, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.-K.; Chiang, M.-C.; Lin, Z.-H.; Lan, G.-Y.; Lin, Y.-W.; Chang, H.-T. Nanomaterial-Based Surface-Assisted Laser Desorption/Ionization Mass Spectrometry of Peptides and Proteins. J. Am. Soc. Mass Spectrom. 2010, 21, 1204–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.; Cheng, Q. Desorption and Ionization Mechanisms and Signal Enhancement in Surface Assisted Laser Desorption Ionization Mass Spectrometry (SALDI-MS). Appl. Spectrosc. Rev. 2020, 55, 220–242. [Google Scholar] [CrossRef]
- Sakai, R.; Ichikawa, T.; Kondo, H.; Ishikawa, K.; Shimizu, N.; Ohta, T.; Hiramatsu, M.; Hori, M. Effects of Carbon Nanowalls (CNWs) Substrates on Soft Ionization of Low-Molecular-Weight Organic Compounds in Surface-Assisted Laser Desorption/Ionization Mass Spectrometry (SALDI-MS). Nanomaterials 2021, 11, 262. [Google Scholar] [CrossRef]
- Kim, S.-W.; Kwon, S.; Kim, Y.-K. Graphene Oxide Derivatives and Their Nanohybrid Structures for Laser Desorption/Ionization Time-of-Flight Mass Spectrometry Analysis of Small Molecules. Nanomaterials 2021, 11, 288. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Q.; Liang, Y.; Jiang, G. Recent Progress in Application of Carbon Nanomaterials in Laser Desorption/Ionization Mass Spectrometry. Anal. Bioanal. Chem. 2016, 408, 2861–2873. [Google Scholar] [CrossRef]
- Najam-ul-Haq, M.; Rainer, M.; Szabó, Z.; Vallant, R.; Huck, C.W.; Bonn, G.K. Role of Carbon Nano-Materials in the Analysis of Biological Materials by Laser Desorption/Ionization-Mass Spectrometry. J. Biochem. Biophys. Methods 2007, 70, 319–328. [Google Scholar] [CrossRef]
- Ohta, T.; Ito, H.; Ishikawa, K.; Kondo, H.; Hiramatsu, M.; Hori, M. Atmospheric Pressure Plasma-Treated Carbon Nanowalls’ Surface-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (CNW-SALDI-MS). C 2019, 5, 40. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Dou, S.; Wang, Z.; Du, J.; Lu, N. Carbon Nanoparticles Derived from Carbon Soot as a Matrix for SALDI-MS Analysis. Microchim. Acta 2020, 187, 161. [Google Scholar] [CrossRef]
- Houdová, D.; Soto, J.; Castro, R.; Rodrigues, J.; Soledad Pino-González, M.; Petković, M.; Bandosz, T.J.; Algarra, M. Chemically Heterogeneous Carbon Dots Enhanced Cholesterol Detection by MALDI TOF Mass Spectrometry. J. Colloid Interface Sci. 2021, 591, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wu, J.; Dong, Y.; Xie, P.; Zhang, Y.; Cai, Z. Nitrogen and Sulfur Co-Doped Carbon-Dot-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry Imaging for Profiling Bisphenol S Distribution in Mouse Tissues. Anal. Chem. 2018, 90, 10872–10880. [Google Scholar] [CrossRef] [PubMed]
- Amini, N.; Shariatgorji, M.; Thorsén, G. SALDI-MS Signal Enhancement Using Oxidized Graphitized Carbon Black Nanoparticles. J. Am. Soc. Mass Spectrom. 2009, 20, 1207–1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, L.; Zheng, G.; Wang, M.; Wang, D.; Xia, Z. Microwave-Prepared Mesoporous Graphene as Adsorbent and Matrix of Surface-Assisted Laser Desorption/Ionization Mass Spectrometry for the Enrichment and Rapid Detection of Polyphenols in Biological Samples. Talanta 2021, 222, 121365. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Arima, K.; Takeshita, T.; Kunitake, Y.; Ohno, N.; Imamura, M.; Matsui, T. Laser Desorption Ionization–Mass Spectrometry with Graphite Carbon Black Nanoparticles for Simultaneous Detection of Taste- and Odor-Active Compounds. ACS Appl. Nano Mater. 2022, 5, 2187–2194. [Google Scholar] [CrossRef]
- Bian, J.; Olesik, S.V. Surface-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry of Small Drug Molecules and High Molecular Weight Synthetic/Biological Polymers Using Electrospun Composite Nanofibers. Analyst 2017, 142, 1125–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.; Song, N.; Dou, S.; Liu, J.; Chen, Q.; Lu, X.; Lu, N. A Flexible SALDI-MS Substrate for No Background Interference Detection. Sens. Actuators B Chem. 2022, 351, 130868. [Google Scholar] [CrossRef]
- Li, F.; Wang, M.; Zhou, J.; Yang, M.; Wang, T. Nanocomposites of Boronic Acid-Functionalized Magnetic Multi-Walled Carbon Nanotubes with Flexible Branched Polymers as a Novel Desorption/Ionization Matrix for the Capture and Direct Detection of Cis-Diol-Flavonoid Compounds Coupled with MALDI-TOF-MS. J. Hazard. Mater. 2022, 429, 128055. [Google Scholar] [CrossRef]
- Zhu, S.; Xu, G. Single-Walled Carbon Nanohorns and Their Applications. Nanoscale 2010, 2, 2538. [Google Scholar] [CrossRef]
- Comisso, N.; Berlouis, L.E.A.; Morrow, J.; Pagura, C. Changes in Hydrogen Storage Properties of Carbon Nano-Horns Submitted to Thermal Oxidation. Int. J. Hydrog. Energy 2010, 35, 9070–9081. [Google Scholar] [CrossRef]
- Calandra, E.; Crotti, S.; Agostini, M.; Nitti, D.; Roverso, M.; Toffoli, G.; Marangon, E.; Posocco, B.; Traldi, P. Matrix-Assisted Laser Desorption/Ionization, Nanostructure-Assisted Laser Desorption/Ionization and Carbon Nanohorns in the Detection of Antineoplastic Drugs. 1. The Cases of Irinotecan, Sunitinib and 6-Alpha-Hydroxy Paclitaxel. Eur. J. Mass Spectrom. 2014, 20, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Lu, M.; Ding, L.; Ju, H.; Cai, Z. Surface-Assisted Laser Desorption/Ionization Mass Spectrometric Detection of Biomolecules by Using Functional Single-Walled Carbon Nanohorns as the Matrix. Chem. Eur. J. 2013, 19, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Meher, J.K.; Dash, G.N.; Meher, P.K.; Raval, M.K. A Reduced Computational Load Protein Coding Predictor Using Equivalent Amino Acid Sequence of DNA String with Period-3 Based Time and Frequency Domain Analysis. AJMB 2011, 1, 79–86. [Google Scholar] [CrossRef] [Green Version]
- Jover, J.; Bosque, R.; Sales, J. A Comparison of the Binding Affinity of the Common Amino Acids with Different Metal Cations. Dalton Trans. 2008, 45, 6441–6453. [Google Scholar] [CrossRef] [PubMed]
- Ackman, R.G.; Sipos, J.C. Application of Specific Response Factors in the Gas Chromatographic Analysis of Methyl Esters of Fatty Acids with Flame Ionization Detectors. J. Am. Oil Chem. Soc. 1964, 41, 377–378. [Google Scholar] [CrossRef]
- Bono, L.; Seraglia, R.; Roverso, M.; Di Carro, M.; Magi, E. Triacylglycerol Profile in Cocoa Liquors Using MALDI-TOF and LC-ESI Tandem Mass Spectrometry: Nine TAGs Identified in Ecuador Cocoa Liquor. J. Mass Spectrom. 2014, 49, 894–899. [Google Scholar] [CrossRef]
- Beltrán, G.; del Rio, C.; Sánchez, S.; Martínez, L. Influence of Harvest Date and Crop Yield on the Fatty Acid Composition of Virgin Olive Oils from Cv. Picual. J. Agric. Food Chem. 2004, 52, 3434–3440. [Google Scholar] [CrossRef]
- Schiavon, M. Device and Method for Production of Carbon Nanotubes, Fullerene and Their Derivatives. U.S. Patent US7125525B2, 24 October 2006. [Google Scholar]








| m/z | Identified FA Composition | Adduct |
|---|---|---|
| 923.498 | 3 × C18:1 (oleic acid) | [M+K]+ |
| 921.463 | 2 × C18:1 (oleic acid) 1 × C18:2 (linoleic acid) | [M+K]+ |
| 907.478 | 3 × C18:1 (oleic acid) | [M+Na]+ |
| 905.435 | 2 × C18:1 (oleic acid) 1 × C18:2 (linoleic acid) | [M+Na]+ |
| 897.479 | 2 × C18:1 (oleic acid) 1 × C16:0 (palmitic acid) | [M+K]+ |
| 895.457 | 1 × C18:1 (oleic acid) 1 × C18:2 (linoleic acid) 1 × C16:0 (palmitic acid) | [M+K]+ |
| 881.455 | 2 × C18:1 (oleic acid) 1 × C16:0 (palmitic acid) | [M+Na]+ |
| 879.489 | 1 × C18:1 (oleic acid) 1 × C18:2 (linoleic acid) 1 × C16:0 (palmitic acid) | [M+Na]+ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roverso, M.; Seraglia, R.; Dogra, R.; Badocco, D.; Pettenuzzo, S.; Cappellin, L.; Pastore, P.; Bogialli, S. Single-Walled Carbon Nanohorns as Boosting Surface for the Analysis of Low-Molecular-Weight Compounds by SALDI-MS. Int. J. Mol. Sci. 2022, 23, 5027. https://doi.org/10.3390/ijms23095027
Roverso M, Seraglia R, Dogra R, Badocco D, Pettenuzzo S, Cappellin L, Pastore P, Bogialli S. Single-Walled Carbon Nanohorns as Boosting Surface for the Analysis of Low-Molecular-Weight Compounds by SALDI-MS. International Journal of Molecular Sciences. 2022; 23(9):5027. https://doi.org/10.3390/ijms23095027
Chicago/Turabian StyleRoverso, Marco, Roberta Seraglia, Raghav Dogra, Denis Badocco, Silvia Pettenuzzo, Luca Cappellin, Paolo Pastore, and Sara Bogialli. 2022. "Single-Walled Carbon Nanohorns as Boosting Surface for the Analysis of Low-Molecular-Weight Compounds by SALDI-MS" International Journal of Molecular Sciences 23, no. 9: 5027. https://doi.org/10.3390/ijms23095027
APA StyleRoverso, M., Seraglia, R., Dogra, R., Badocco, D., Pettenuzzo, S., Cappellin, L., Pastore, P., & Bogialli, S. (2022). Single-Walled Carbon Nanohorns as Boosting Surface for the Analysis of Low-Molecular-Weight Compounds by SALDI-MS. International Journal of Molecular Sciences, 23(9), 5027. https://doi.org/10.3390/ijms23095027