Orientational Preferences of GPI-Anchored Ly6/uPAR Proteins
Abstract
:1. Introduction
2. Results and Discussion
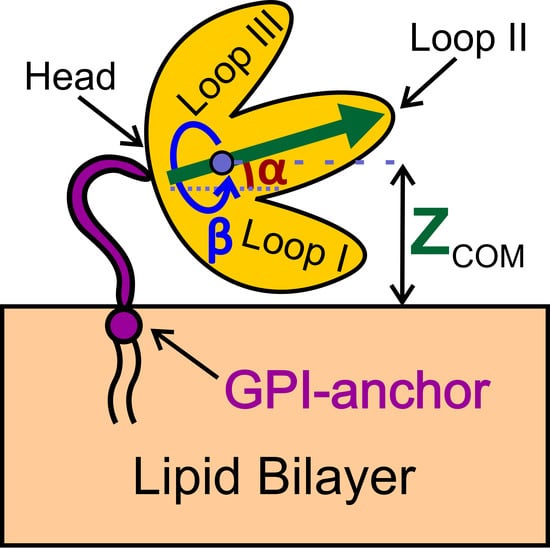
2.1. Centers of Mass (COM) Positions
2.2. Orientational Preferences
2.3. Interaction of Ly6 Proteins with Membrane Lipids
2.4. Data Relevance and Application to In Vivo
3. Materials and Methods
3.1. Systems Preparation
3.2. Molecular Dynamics Simulations
3.3. Data Analysis
3.4. Lipid Contacts
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vasilyeva, N.A.; Loktyushov, E.V.; Bychkov, M.L.; Shenkarev, Z.O.; Lyukmanova, E.N. Three-Finger Proteins from the Ly6/uPAR Family: Functional Diversity within One Structural Motif. Biochemistry 2017, 82, 1702–1715. [Google Scholar] [CrossRef] [PubMed]
- Leth, J.M.; Leth-Espensen, K.Z.; Kristensen, K.K.; Kumari, A.; Lund Winther, A.-M.; Young, S.G.; Ploug, M. Evolution and Medical Significance of LU Domain-Containing Proteins. Int. J. Mol. Sci. 2019, 20, 2760. [Google Scholar] [CrossRef] [Green Version]
- Miwa, J.M. Lynx1 prototoxins: Critical accessory proteins of neuronal nicotinic acetylcholine receptors. Curr. Opin. Pharmacol. 2021, 56, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Shabelnikov, S.V.; Bobkov, D.E.; Sharlaimova, N.S.; Petukhova, O.A. Injury affects coelomic fluid proteome of the common starfish, Asterias rubens. J. Exp. Biol. 2019, 222, jeb198556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paramonov, A.S.; Shulepko, M.A.; Makhonin, A.M.; Bychkov, M.L.; Kulbatskii, D.S.; Chernikov, A.M.; Myshkin, M.Y.; Shabelnikov, S.V.; Shenkarev, Z.O.; Kirpichnikov, M.P.; et al. New Three-Finger Protein from Starfish Asteria rubens Shares Structure and Pharmacology with Human Brain Neuromodulator Lynx2. Mar. Drugs 2022, 20, 503. [Google Scholar] [CrossRef]
- Davey, S.D.; Chalmers, I.W.; Fernandez-Fuentes, N.; Swain, M.T.; Smith, D.; Abbas Abidi, S.M.; Saifullah, M.K.; Raman, M.; Ravikumar, G.; McVeigh, P.; et al. In silico characterisation of the complete Ly6 protein family in Fasciola gigantica supported through transcriptomics of the newly-excysted juveniles. Mol. Omics 2022, 18, 45–56. [Google Scholar] [CrossRef]
- Koh, K.; Joiner, W.J.; Wu, M.N.; Yue, Z.; Smith, C.J.; Sehgal, A. Identification of SLEEPLESS, a sleep-promoting factor. Science 2008, 321, 372–376. [Google Scholar] [CrossRef] [Green Version]
- Upadhyay, G. Emerging Role of Lymphocyte Antigen-6 Family of Genes in Cancer and Immune Cells. Front. Immunol. 2019, 10, 819. [Google Scholar] [CrossRef]
- Zhai, B.-T.; Tian, H.; Sun, J.; Zou, J.-B.; Zhang, X.-F.; Cheng, J.-X.; Shi, Y.-J.; Fan, Y.; Guo, D.-Y. Urokinase-type plasminogen activator receptor (uPAR) as a therapeutic target in cancer. J. Transl. Med. 2022, 20, 135. [Google Scholar] [CrossRef]
- Loughner, C.L.; Bruford, E.A.; McAndrews, M.S.; Delp, E.E.; Swamynathan, S.; Swamynathan, S.K. Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Hum. Genom. 2016, 10, 10. [Google Scholar] [CrossRef]
- Paramonov, A.S.; Kocharovskaya, M.V.; Tsarev, A.V.; Kulbatskii, D.S.; Loktyushov, E.V.; Shulepko, M.A.; Kirpichnikov, M.P.; Lyukmanova, E.N.; Shenkarev, Z.O. Structural Diversity and Dynamics of Human Three-Finger Proteins Acting on Nicotinic Acetylcholine Receptors. Int. J. Mol. Sci. 2020, 21, 7280. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M.; Koh, C.Y. Snake venom three-finger toxins and their potential in drug development targeting cardiovascular diseases. Biochem. Pharmacol. 2020, 181, 114105. [Google Scholar] [CrossRef] [PubMed]
- Kessler, P.; Marchot, P.; Silva, M.; Servent, D. The three-finger toxin fold: A multifunctional structural scaffold able to modulate cholinergic functions. J. Neurochem. 2017, 142 (Suppl. 2), 7–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Utkin, Y.N. Last decade update for three-finger toxins: Newly emerging structures and biological activities. World J. Biol. Chem. 2019, 10, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Campos, P.C.; de Melo, L.A.; Dias, G.L.F.; Fortes-Dias, C.L. Endogenous phospholipase A2 inhibitors in snakes: A brief overview. J. Venom. Anim. Toxins Incl. Trop. Dis. 2016, 22, 37. [Google Scholar] [CrossRef] [Green Version]
- Anderson, K.R.; Hoffman, K.M.; Miwa, J.M. Modulation of cholinergic activity through lynx prototoxins: Implications for cognition and anxiety regulation. Neuropharmacology 2020, 174, 108071. [Google Scholar] [CrossRef]
- Shulepko, M.A.; Bychkov, M.L.; Shenkarev, Z.O.; Kulbatskii, D.S.; Makhonin, A.M.; Paramonov, A.S.; Chugunov, A.O.; Kirpichnikov, M.P.; Lyukmanova, E.N. Biochemical Basis of Skin Disease Mal de Meleda: SLURP-1 Mutants Differently Affect Keratinocyte Proliferation and Apoptosis. J. Investig. Dermatol. 2021, 141, 2229–2237. [Google Scholar] [CrossRef]
- Lyukmanova, E.N.; Shulepko, M.A.; Shenkarev, Z.O.; Bychkov, M.L.; Paramonov, A.S.; Chugunov, A.O.; Kulbatskii, D.S.; Arvaniti, M.; Dolejsi, E.; Schaer, T.; et al. Secreted Isoform of Human Lynx1 (SLURP-2): Spatial Structure and Pharmacology of Interactions with Different Types of Acetylcholine Receptors. Sci. Rep. 2016, 6, 30698. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, Q.; Liao, Q.; Zhao, Y. CD59: A promising target for tumor immunotherapy. Future Oncol. 2018, 14, 781–791. [Google Scholar] [CrossRef]
- Huang, Y.; Smith, C.A.; Song, H.; Morgan, B.P.; Abagyan, R.; Tomlinson, S. Insights into the human CD59 complement binding interface toward engineering new therapeutics. J. Biol. Chem. 2005, 280, 34073–34079. [Google Scholar] [CrossRef]
- Lyukmanova, E.N.; Shenkarev, Z.O.; Shulepko, M.A.; Mineev, K.S.; D’Hoedt, D.; Kasheverov, I.E.; Filkin, S.Y.; Krivolapova, A.P.; Janickova, H.; Dolezal, V.; et al. NMR structure and action on nicotinic acetylcholine receptors of water-soluble domain of human LYNX1. J. Biol. Chem. 2011, 286, 10618–10627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Özhan, G.; Sezgin, E.; Wehner, D.; Pfister, A.S.; Kühl, S.J.; Kagermeier-Schenk, B.; Kühl, M.; Schwille, P.; Weidinger, G. Lypd6 enhances Wnt/β-catenin signaling by promoting Lrp6 phosphorylation in raft plasma membrane domains. Dev. Cell 2013, 26, 331–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Ren, J.; Lu, W.; Harlos, K.; Jones, E.Y. Structure of the Wnt signaling enhancer LYPD6 and its interactions with the Wnt coreceptor LRP6. FEBS Lett. 2018, 592, 3152–3162. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Kern, N.R.; Anderson, K.R.; Zhang, X.F.; Miwa, J.M.; Im, W. Dynamics and Interactions of GPI-Linked lynx1 Protein with/without Nicotinic Acetylcholine Receptor in Membrane Bilayers. J. Phys. Chem. B 2020, 124, 4017–4025. [Google Scholar] [CrossRef] [PubMed]
- Shenkarev, Z.O.; Shulepko, M.A.; Bychkov, M.L.; Kulbatskii, D.S.; Shlepova, O.V.; Vasilyeva, N.A.; Andreev-Andrievskiy, A.A.; Popova, A.S.; Lagereva, E.A.; Loktyushov, E.V.; et al. Water-soluble variant of human Lynx1 positively modulates synaptic plasticity and ameliorates cognitive impairment associated with α7-nAChR dysfunction. J. Neurochem. 2020, 155, 45–61. [Google Scholar] [CrossRef]
- Miwa, J.M.; Stevens, T.R.; King, S.L.; Caldarone, B.J.; Ibanez-Tallon, I.; Xiao, C.; Fitzsimonds, R.M.; Pavlides, C.; Lester, H.A.; Picciotto, M.R.; et al. The prototoxin lynx1 acts on nicotinic acetylcholine receptors to balance neuronal activity and survival in vivo. Neuron 2006, 51, 587–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morishita, H.; Miwa, J.M.; Heintz, N.; Hensch, T.K. Lynx1, a cholinergic brake, limits plasticity in adult visual cortex. Science 2010, 330, 1238–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horie, M.; Okutomi, K.; Taniguchi, Y.; Ohbuchi, Y.; Suzuki, M.; Takahashi, E.-I. Isolation and characterization of a new member of the HumanLy6Gene family(LY6H). Genomics 1998, 53, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, Y.; Kubo, N.; Watanabe, M.; Asano, S.; Shinoda, T.; Sugino, T.; Ichikawa, D.; Tsuji, S.; Kato, F.; Misawa, H. Endogenous neurotoxin-like protein Ly6H inhibits alpha7 nicotinic acetylcholine receptor currents at the plasma membrane. Sci. Rep. 2020, 10, 11996. [Google Scholar] [CrossRef]
- Wu, M.; Liu, C.Z.; Barrall, E.A.; Rissman, R.A.; Joiner, W.J. Unbalanced regulation of α7 nAChRs by Ly6h and NACHO contributes to neurotoxicity in Alzheimer’s disease. J. Neurosci. 2021, 41, 8461–8474. [Google Scholar] [CrossRef]
- Dessaud, E.; Salaün, D.; Gayet, O.; Chabbert, M.; de Lapeyrière, O. Identification of lynx2, a novel member of the ly-6/neurotoxin superfamily, expressed in neuronal subpopulations during mouse development. Mol. Cell. Neurosci. 2006, 31, 232–242. [Google Scholar] [CrossRef]
- Paramonov, A.S.; Shulepko, M.A.; Kocharovskaya, M.V.; Alenkin, A.E.; Evdokimova, A.O.; Akentiev, P.I.; Shenkarev, Z.O.; Kirpichnikov, M.P.; Lyukmanova, E.N. Bacterial Production and Structural Study of Human Neuromodulator Lynx2. Russ. J. Bioorganic Chem. 2020, 46, 1261–1269. [Google Scholar] [CrossRef]
- Tekinay, A.B.; Nong, Y.; Miwa, J.M.; Lieberam, I.; Ibanez-Tallon, I.; Greengard, P.; Heintz, N. A role for LYNX2 in anxiety-related behavior. Proc. Natl. Acad. Sci. USA 2009, 106, 4477–4482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Lang, Q.; Li, J.; Xie, F.; Wan, B.; Yu, L. Identification and characterization of human LYPD6, a new member of the Ly-6 superfamily. Mol. Biol. Rep. 2010, 37, 2055–2062. [Google Scholar] [CrossRef] [PubMed]
- Kulbatskii, D.; Shenkarev, Z.; Bychkov, M.; Loktyushov, E.; Shulepko, M.; Koshelev, S.; Povarov, I.; Popov, A.; Peigneur, S.; Chugunov, A.; et al. Human Three-Finger Protein Lypd6 Is a Negative Modulator of the Cholinergic System in the Brain. Front. Cell Dev. Biol. 2021, 9, 2593. [Google Scholar] [CrossRef]
- Sadahiro, M.; Demars, M.P.; Burman, P.; Yevoo, P.; Zimmer, A.; Morishita, H. Activation of somatostatin interneurons by nicotinic modulator Lypd6 enhances plasticity and functional recovery in the adult mouse visual cortex. J. Neurosci. 2020, 40, 5214–5227. [Google Scholar] [CrossRef]
- Arvaniti, M.; Polli, F.S.; Kohlmeier, K.A.; Thomsen, M.S.; Andreasen, J.T. Loss of Lypd6 leads to reduced anxiety-like behaviour and enhanced responses to nicotine. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 82, 86–94. [Google Scholar] [CrossRef]
- Ni, J.; Lang, Q.; Bai, M.; Zhong, C.; Chen, X.; Wan, B.; Yu, L. Cloning and characterization of a human LYPD7, a new member of the Ly-6 superfamily. Mol. Biol. Rep. 2009, 36, 697–703. [Google Scholar] [CrossRef]
- Paramonov, A.S.; Kulbatskii, D.S.; Loktyushov, E.V.; Tsarev, A.V.; Dolgikh, D.A.; Shenkarev, Z.O.; Kirpichnikov, M.P.; Lyukmanova, E.N. Recombinant production and structural studies of the human Lypd6 and Lypd6b proteins. Russ. J. Bioorganic Chem. 2017, 43, 644–652. [Google Scholar] [CrossRef]
- Ochoa, V.; George, A.A.; Nishi, R.; Whiteaker, P. The prototoxin LYPD6B modulates heteromeric α3β4-containing nicotinic acetylcholine receptors, but not α7 homomers. FASEB J. 2016, 30, 1109–1119. [Google Scholar] [CrossRef]
- Morgan, P. Chapter 34—CD59. In The Complement FactsBook, 2nd ed.; Barnum, S., Schein, T., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 361–367. [Google Scholar]
- Li, I.T.S.; Walker, G.C. Signature of hydrophobic hydration in a single polymer. Proc. Natl. Acad. Sci. USA 2011, 108, 16527–16532. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, P.; Wehle, M.; Lipowsky, R.; Santer, M. A molecular dynamics model for glycosylphosphatidyl-inositol anchors: “flop down” or “lollipop”? Phys. Chem. Chem. Phys. 2018, 20, 29314–29324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langelaan, D.N.; Rainey, J.K. Membrane catalysis of peptide-receptor binding. Biochem. Cell Biol. 2010, 88, 203–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castanho, M.A.R.B.; Fernandes, M.X. Lipid membrane-induced optimization for ligand-receptor docking: Recent tools and insights for the “membrane catalysis” model. Eur. Biophys. J. 2006, 35, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Lyukmanova, E.N.; Shulepko, M.A.; Buldakova, S.L.; Kasheverov, I.E.; Shenkarev, Z.O.; Reshetnikov, R.V.; Filkin, S.Y.; Kudryavtsev, D.S.; Ojomoko, L.O.; Kryukova, E.V.; et al. Water-soluble LYNX1 residues important for interaction with muscle-type and/or neuronal nicotinic receptors. J. Biol. Chem. 2013, 288, 15888–15899. [Google Scholar] [CrossRef] [Green Version]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Patel, D.S.; Ståhle, J.; Park, S.-J.; Kern, N.R.; Kim, S.; Lee, J.; Cheng, X.; Valvano, M.A.; Holst, O.; et al. CHARMM-GUI Membrane Builder for Complex Biological Membrane Simulations with Glycolipids and Lipoglycans. J. Chem. Theory Comput. 2019, 15, 775–786. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-J.; Lee, J.; Qi, Y.; Kern, N.R.; Lee, H.S.; Jo, S.; Joung, I.; Joo, K.; Lee, J.; Im, W. CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates. Glycobiology 2019, 29, 320–331. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Søndergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P.M.; Morgan, B.P.; Wormald, M.R.; Harvey, D.J.; van den Berg, C.W.; Davis, S.J.; Ferguson, M.A.; Dwek, R.A. The glycosylation of the complement regulatory protein, human erythrocyte CD59. J. Biol. Chem. 1997, 272, 7229–7244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudd, P.M.; Morgan, B.P.; Wormald, M.R.; Harvey, D.J.; van den Berg, C.W.; Davis, S.J.; Ferguson, M.A.; Dwek, R.A. Roles for glycosylation in the anti-inflammatory molecule CD59. Biochem. Soc. Trans. 1997, 25, 1177–1184. [Google Scholar] [CrossRef] [Green Version]
- Meri, S.; Lehto, T.; Sutton, C.W.; Tyynelä, J.; Baumann, M. Structural composition and functional characterization of soluble CD59: Heterogeneity of the oligosaccharide and glycophosphoinositol (GPI) anchor revealed by laser-desorption mass spectrometric analysis. Biochem. J. 1996, 316 Pt 3, 923–935. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, C.M.; Harrison, R.A.; Lachmann, P.J.; Neuhaus, D. Structure of a soluble, glycosylated form of the human complement regulatory protein CD59. Structure 1994, 2, 185–199. [Google Scholar] [CrossRef] [Green Version]
- Barboza, M.; Solakyildirim, K.; Knotts, T.A.; Luke, J.; Gareau, M.G.; Raybould, H.E.; Lebrilla, C.B. Region-Specific Cell Membrane N-Glycome of Functional Mouse Brain Areas Revealed by nanoLC-MS Analysis. Mol. Cell. Proteom. 2021, 20, 100130. [Google Scholar] [CrossRef]
- Kinoshita, T. Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol. 2020, 10, 190290. [Google Scholar] [CrossRef] [Green Version]
- Bernecker, C.; Köfeler, H.; Pabst, G.; Trötzmüller, M.; Kolb, D.; Strohmayer, K.; Trajanoski, S.; Holzapfel, G.A.; Schlenke, P.; Dorn, I. Cholesterol Deficiency Causes Impaired Osmotic Stability of Cultured Red Blood Cells. Front. Physiol. 2019, 10, 1529. [Google Scholar] [CrossRef]
- Sunshine, C.; McNamee, M.G. Lipid modulation of nicotinic acetylcholine receptor function: The role of membrane lipid composition and fluidity. Biochim. Biophys. Acta 1994, 1191, 59–64. [Google Scholar] [CrossRef]
- Guvench, O.; Mallajosyula, S.S.; Raman, E.P.; Hatcher, E.; Vanommeslaeghe, K.; Foster, T.J.; Jamison, F.W., 2nd; Mackerell, A.D., Jr. CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comput. 2011, 7, 3162–3180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallajosyula, S.S.; Jo, S.; Im, W.; MacKerell, A.D., Jr. Molecular dynamics simulations of glycoproteins using CHARMM. Methods Mol. Biol. 2015, 1273, 407–429. [Google Scholar] [PubMed] [Green Version]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Khan, H.M.; MacKerell, A.D., Jr.; Reuter, N. Cation-π Interactions between Methylated Ammonium Groups and Tryptophan in the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2019, 15, 7–12. [Google Scholar] [CrossRef]
- Bernetti, M.; Bussi, G. Pressure control using stochastic cell rescaling. J. Chem. Phys. 2020, 153, 114107. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Procter, J.B.; Carstairs, G.M.; Soares, B.; Mourão, K.; Ofoegbu, T.C.; Barton, D.; Lui, L.; Menard, A.; Sherstnev, N.; Roldan-Martinez, D.; et al. Alignment of Biological Sequences with Jalview. Methods Mol. Biol. 2021, 2231, 203–224. [Google Scholar]
- Shafee, T.M.A.; Robinson, A.J.; van der Weerden, N.; Anderson, M.A. Structural homology guided alignment of cysteine rich proteins. Springerplus 2016, 5, 27. [Google Scholar] [CrossRef] [Green Version]
- Krylov, N.A.; Efremov, R.G. libxtc: An efficient library for reading XTC-compressed MD trajectory data. BMC Res. Notes 2021, 14, 124. [Google Scholar] [CrossRef] [PubMed]
- Pyrkov, T.V.; Chugunov, A.O.; Krylov, N.A.; Nolde, D.E.; Efremov, R.G. PLATINUM: A web tool for analysis of hydrophobic/hydrophilic organization of biomolecular complexes. Bioinformatics 2009, 25, 1201–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Protein | Mature Form, Residues a | Glycosylation Sites | N-Terminal Region Length, Residues b | C-Terminal Linker Length, Residues c | COM Position, nm d | Tilt Angle α, ° d | Rotation Angle β, ° d |
|---|---|---|---|---|---|---|---|
| Lynx1 | 21–91 | — | 0 | 0 | 1.43 ± 0.21 | –31.5 ± 8.7 (0.7 ± 6.5) | −121.6 ± 8.6 −47.9 ± 7.2 |
| Lynx2 | 21–117 | N: Asn45 | 2 | 11 | 1.93 ± 0.23 | −43.8 ± 5.6 (−18.8 ± 37.8) e (40.9 ± 4.6) | −37.9 ± 5.8 (20.4 ± 4.4) (22.0 ± 38.1) e |
| Lypd6 | 23–147 | N: Asn134 | 24 | 20 | 1.62 ± 0.20 (2.23 ± 0.20) | 17.3 ± 4.4 31.7 ± 5.9 (60.1 ± 12.3) | −139.9 ± 4.2 −146.2 ± 15.4 (166.6 ± 10.9) |
| Lypd6B | 40–164 | N: Asn147 | 20 | 24 | 2.00 ± 0.16 | −31.5 ± 9.4 | −122.6 ± 9.4 −57.0 ± 8.9 |
| Ly6H | 26–115 | N: Asn36 | 0 | 5 | 1.78 ± 0.20 2.27 ± 0.18 | −46.6 ± 12.0 (37.6 ± 16.1) | −15.7 ± 11.1 −144.0 ± 9.2 |
| CD59 | 26–102 | O: Thr76, Thr77 N: Asn43 | 0 | 8 | 1.29 ± 0.13 | 17.8 ± 9.0 | 29.8 ± 7.9 |
| Protein | Ion–Ion and Ion–Dipole Interactions | Hydrogen Bonds | π–Cation Interactions | Hydrophobic Contacts |
|---|---|---|---|---|
| Lynx1 | Loop I: D31, R38, Loop II: R57, K59 GPI: DSPI-1, GlcN-2, PEtN-Man-3 | Loop I: Y28, N29, G30, N32, C33, F34, N35 Loop II: Y53, T54, T56 Loop III: Y76 | — | Loop I: C26, A27, P36 Loop II: P55 |
| Lynx2 | Loop I: E30 Loop III: K92 C-terminus: R110, K112, K113, R114 GPI: DSPI-1, GlcN-2, PEtN-Man-3 | Loop I: Q32 Loop III: Y84 C-terminus: G115, S116 N-glycan: GlcNAc-8 | Loop I: F31 | Loop I: N35 Loop II: A63 |
| Lypd6 | N-terminus: R26, K31 Loop I: R63, D67 Head I: R72, E73 Head II: R75 GPI: DSPI-1, GlcN-2, PEtN-Man-3 | N-terminus: Y43, G46 Loop I: W64, Y69 GPI: Man-6 | Loop I: W64, Y69 | N-terminus: P44, G45, K48 Loop I: P66, I68 Head I: P71 GPI: Man-5 |
| Lypd6B | Loop I: R76 Loop II: R101 GPI: DSPI-1, GlcN-2, PetN-Man-3 | N-terminus: Y45, N46 Loop I: Y72, N73, W77 N-glycan: Man-4, GlcNAc-6, GlcNAc-8 | Loop I: W77 | N-terminus: N43, V47, P49, P50 Loop I: D70 C-terminus: S164 GPI: Man-5 |
| Ly6H | Loop II: R64, K65 Head III: K107 GPI: DSPI-1, GlcN-2, PEtN-Man-3 | Loop I: T35 Loop II: S63 Loop III: Y88 N-glycan: GlcNAc-5, Man-7, GlcNAc-8 GPI: Man-4, Man-5 | Loop I: H39 | Loop III: F92 N-glycan: GlcNAc-1 |
| CD59 | Head III: K90, K91 C-terminus: E101 GPI: GlcN-2, PEtN-Man-3 | Loop III: Y86, Y87 C-terminus: Q99, N102 O-glycan (T76): Neu5Ac-3 GPI: SAPI-1 | Head II: F67 | Head II: E68, C70 Loop III: N71, F72, N73 Head III: C88, C89, C94 C-terminus: F96, E98, L100 O-glycan (T76): Gal-2 GPI: Man-5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaigraev, M.M.; Lyukmanova, E.N.; Paramonov, A.S.; Shenkarev, Z.O.; Chugunov, A.O. Orientational Preferences of GPI-Anchored Ly6/uPAR Proteins. Int. J. Mol. Sci. 2023, 24, 11. https://doi.org/10.3390/ijms24010011
Zaigraev MM, Lyukmanova EN, Paramonov AS, Shenkarev ZO, Chugunov AO. Orientational Preferences of GPI-Anchored Ly6/uPAR Proteins. International Journal of Molecular Sciences. 2023; 24(1):11. https://doi.org/10.3390/ijms24010011
Chicago/Turabian StyleZaigraev, Maxim M., Ekaterina N. Lyukmanova, Alexander S. Paramonov, Zakhar O. Shenkarev, and Anton O. Chugunov. 2023. "Orientational Preferences of GPI-Anchored Ly6/uPAR Proteins" International Journal of Molecular Sciences 24, no. 1: 11. https://doi.org/10.3390/ijms24010011
APA StyleZaigraev, M. M., Lyukmanova, E. N., Paramonov, A. S., Shenkarev, Z. O., & Chugunov, A. O. (2023). Orientational Preferences of GPI-Anchored Ly6/uPAR Proteins. International Journal of Molecular Sciences, 24(1), 11. https://doi.org/10.3390/ijms24010011