RNAi Technology: A New Path for the Research and Management of Obligate Biotrophic Phytopathogenic Fungi
Abstract
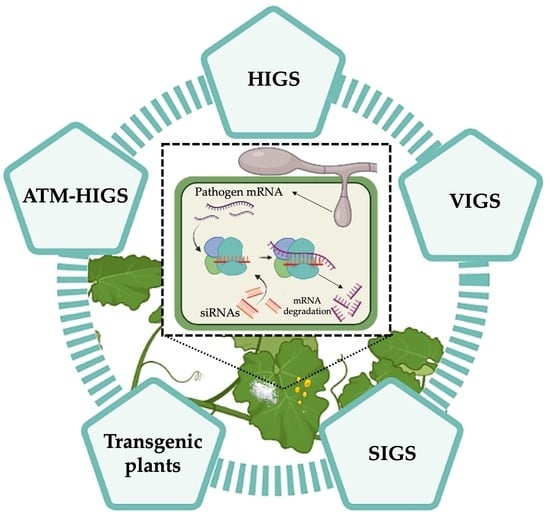
:1. Introduction
2. Powdery Mildew and Rust Fungi
3. RNAi Tools for Gene Function Analysis of Obligate Biotrophic Fungi
3.1. Virus-Induced Gene Silencing (VIGS)
3.2. Host Induced Gene Silencing (HIGS)
3.3. Agrobacterium tumefaciens-Mediated Host-Induced Gene Silencing (ATM-HIGS)
3.4. Direct Application of dsRNA
4. Control of Powdery Mildew and Rust Diseases by RNAi Technology
4.1. Transgenic Plants Expressing RNAi Constructs
| Plant Host | Cultivar | Pathogen | Target Gene | Gene Function | Effects | References |
|---|---|---|---|---|---|---|
| H. vulgare | Golden Promise | Blumeria graminis | BgGTF1 | 1,3-β-glucanosyltransferase 1 | Reduced manifestation of powdery mildew symptoms | [53] |
| T. aestivum | Bobwhite | B. graminis f. sp. tritici | SvrPm3a1/f1 | RNase-like effector | Enhanced resistance to powdery mildew | [9] |
| Bgt-Bcg-6 | ||||||
| Bgt-Bcg-7 | ||||||
| Xinong1376 | Puccinia striiformis f. sp. tritici | PsFUZ7 | MAPK kinase | Enhanced resistance to rust | [68] | |
| PKA | Protein kinase A | Enhanced resistance to rust | [70] | |||
| PsCPK1 | Catalytic subunit | |||||
| Fielder | Pst_4 | Effector | Enhanced resistance to rust | [111] | ||
| Pst_5 | ||||||
| Fielder | Puccinia triticina | PtMAPK1 | MAP kinase | Reduction of wheat leaf rust disease symptoms | [111] | |
| PtCYC1 | Cyclophilin |
4.2. Spray-Induced Gene Silencing (SIGS)
| Plant Host | Cultivar | Pathogen | Target Gene | Possible Gene Function | RNA Amount | RNA Application | Effects | References |
|---|---|---|---|---|---|---|---|---|
| C. melo | cv. Rochet | Podosphaera xanthii | PxCNAP1048 | Glycosylation | 5–30 μg/mL | Leaves were spray-inoculated with 104 conidia/mL after dsRNA application | Effective management of PM disease | [29] |
| PxCNAP10905 | Respiration | |||||||
| PxCNAP30520 | ||||||||
| G. max | cv. Enrei | Phakopsora pachyrhizi | ATC | Acetyl-CoA acyltransferase | 20 μg/mL | Leaves were spray-inoculated with 105 uredinia/mL after dsRNA application | Effective management of Asian soybean rust (ASR) disease | [115] |
| RP_S16 | 40S ribosomal protein S16 | |||||||
| GCS_H | Glycine cleavage system H protein | |||||||
| CHS | Chitin synthase | 10 ng/mL | Leaves were drop-inoculated with 105 uredinia/mL and dsRNA simultaneously | Effective management of Asian soybean rust (ASR) disease | [100] | |||
| Syzygium jambos | - | Austropuccinia psidii | β-TUB | β-tubulin | 100 ng/μL | Young, emerging leaves were inoculated with 1 mL of dsRNA solutions | Reduction in fungal growth and in the number of urediniospores | [116] |
| EF1-a | Translation elongation factor 1ɑ | |||||||
| ATC | Acetyl-CoA transferase | |||||||
| CYP450 | Cytochrome P450 | |||||||
| MAPK | Mitogen-activated protein kinase | |||||||
| GCS-H | Glycine cleavage system H | |||||||
| 28S rRNA | 28S ribosomal RNA | |||||||
| HAUS01215 | Haustoria target |
5. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gheysen, G.; Vanholme, B. RNAi from plants to nematodes. Trends Biotechnol. 2007, 25, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008, 23, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed]
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big impact on plant development. Trends Plant Sci. 2017, 22, 1056–1068. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, J.; Tung, J.; Liu, D.; Zhou, Y.; He, S.; Du, Y.; Baker, B.; Li, F. A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 2018, 14, e1006756. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.; Zhao, J.H.; Guo, H.S. Trans-kingdom RNA silencing in plant–fungal pathogen interactions. Mol. Plant 2018, 11, 235–244. [Google Scholar] [CrossRef]
- Rosa, C.; Kuo, Y.W.; Wuriyanghan, H.; Falk, B.W. RNA interference mechanisms and applications in plant pathology. Annu. Rev. Phytopathol. 2018, 56, 581–610. [Google Scholar] [CrossRef]
- Muhammad, T.; Zhang, F.; Zhang, Y.; Liang, Y. RNA interference: A natural immune system of plants to counteract biotic stressors. Cells 2019, 8, 38. [Google Scholar] [CrossRef]
- Schaefer, L.K.; Parlange, F.; Buchmann, G.; Jung, E.; Wehrli, A.; Herren, G.; Müller, M.C.; Stehlin, J.; Schmid, R.; Wicker, T.; et al. Cross-kingdom RNAi of pathogen effectors leads to quantitative adult plant resistance in wheat. Front. Plant Sci. 2020, 11, 253. [Google Scholar] [CrossRef]
- Zrachya, A.; Kumar, P.P.; Ramakrishnan, U.; Levy, Y.; Loyter, A.; Arazi, T.; Lapidot, M.; Gafni, Y. Production of siRNA targeted against TYLCV coat protein transcripts leads to silencing of its expression and resistance to the virus. Transgenic Res. 2007, 16, 385–398. [Google Scholar] [CrossRef]
- Puyam, A.; Sharma, S.; Kashyap, P.L. RNA interference- a novel approach for plant disease management. J. Appl. Nat. Sci. 2017, 9, 1612–1618. [Google Scholar] [CrossRef]
- Vaucheret, H.; Vazquez, F.; Crété, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Limera, C.; Sabbadini, S.; Sweet, J.B.; Mezzetti, B. New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 2017, 8, 1418. [Google Scholar] [CrossRef] [PubMed]
- Gebremichael, D.E.; Haile, Z.M.; Negrini, F.; Sabbadini, S.; Capriotti, L.; Mezzetti, B.; Baraldi, E. RNA interference strategies for future management of plant pathogenic fungi: Prospects and challenges. Plants 2021, 10, 650. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Rossi, J. RNAi mechanisms and applications. BioTechniques 2008, 44, 613–616. [Google Scholar] [CrossRef]
- Knip, M.; Constantin, M.E.; Thordal-Christensen, H. Trans-kingdom cross-talk: Small RNAs on the move. PLoS Genet. 2014, 10, e1004602. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Mu, J.; Zhuang, X.; Wang, Q.; Jiang, H.; Deng, Z.B.; Wang, B.; Zhang, L.; Kakar, S.; Jun, Y.; Miller, D.; et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014, 58, 1561–1573. [Google Scholar] [CrossRef]
- Raimondo, S.; Naselli, F.; Fontana, S.; Monteleone, F.; Lo Dico, A.; Saieva, L.; Zito, G.; Flugy, A.; Manno, M.; Di Bella, M.A.; et al. Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget 2015, 6, 19514–19527. [Google Scholar] [CrossRef]
- Wytinck, N.; Manchur, C.L.; Li, V.H.; Whyard, S.; Belmonte, M.F. dsRNA uptake in plant pests and pathogens: Insights into RNAi-based insect and fungal control technology. Plants 2020, 9, 1780. [Google Scholar] [CrossRef]
- Shih, J.D.; Hunter, C.P. SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 2011, 17, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Aizawa, S.; Fujiwara, Y.; Contu, V.R.; Hase, K.; Takahashi, M.; Kikuchi, H.; Kabuta, C.; Wada, K.; Kabuta, T. Lysosomal putative RNA transporter SIDT2 mediates direct uptake of RNA by lysosomes. Autophagy 2016, 12, 565–578. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weiberg, A.; Lin, F.-M.; Thomma, B.P.H.J.; Huang, H.-D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef]
- McLoughlin, A.G.; Wytinck, N.; Walker, P.L.; Girard, I.J.; Rashid, K.Y.; de Kievit, T.; Fernando, W.G.D.; Whyard, S.; Belmonte, M.F. Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea. Sci. Rep. 2018, 8, 7320. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Sandrini, M.; Gambino, G.; Chitarra, W. Double-stranded RNAs (dsRNAs) as a sustainable tool against gray mold (Botrytis cinerea) in grapevine: Effectiveness of different application methods in an open-air environment. Biomolecules 2020, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog 2016, 12, e1005901. [Google Scholar] [CrossRef]
- Werner, B.T.; Gaffar, F.Y.; Schuemann, J.; Biedenkopf, D.; Koch, A.M. RNA-spray-mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance. Front. Plant Sci. 2020, 11, 476. [Google Scholar] [CrossRef]
- Marcianò, D.; Ricciardi, V.; Marone Fassolo, E.; Passera, A.; Bianco, P.A.; Failla, O.; Casati, P.; Maddalena, G.; De Lorenzis, G.; Toffolatti, S.L. RNAi of a putative grapevine susceptibility gene as a possible downy mildew control strategy. Front. Plant Sci. 2021, 12, 667319. [Google Scholar] [CrossRef]
- Ruiz-Jiménez, L.; Polonio, Á.; Vielba-Fernández, A.; Pérez-García, A.; Fernández-Ortuño, D. Gene mining for conserved, non-annotated proteins of Podosphaera xanthii identifies novel target candidates for controlling powdery mildews by spray-induced gene silencing. J Fungi 2021, 7, 735. [Google Scholar] [CrossRef]
- Fitzgerald, A.; van Kan, J.A.L.; Plummer, K.M. Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet. Biol. 2004, 41, 963–971. [Google Scholar] [CrossRef]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef]
- Koeck, M.; Hardham, A.R.; Dodds, P.N. The role of effectors of biotrophic and hemibiotrophic fungi in infection: Effectors of biotrophic fungi. Cell. Microbiol. 2011, 13, 1849–1857. [Google Scholar] [CrossRef]
- Tang, C.; Xu, Q.; Zhao, M.; Wang, X.; Kang, Z. Understanding the lifestyles and pathogenicity mechanisms of obligate biotrophic fungi in wheat: The emerging genomics era. Crop. J. 2018, 6, 60–67. [Google Scholar] [CrossRef]
- Braun, U. The current systematics and taxonomy of the powdery mildews (Erysiphales): An overview. Mycoscience 2011, 52, 210–212. [Google Scholar] [CrossRef]
- Vielba-Fernández, A.; Polonio, Á.; Ruiz-Jiménez, L.; de Vicente, A.; Pérez-García, A.; Fernández-Ortuño, D. Fungicide resistance in powdery mildew fungi. Microorganisms 2020, 8, 1431. [Google Scholar] [CrossRef]
- Pérez-García, A.; Romero, D.; Fernández-Ortuño, D.; López-Ruiz, F.; De Vicente, A.; Torés, J.A. The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol. Plant Pathol. 2009, 10, 153–160. [Google Scholar] [CrossRef]
- Eichmann, R.; Hückelhoven, R. Accommodation of powdery mildew fungi in intact plant cells. J. Plant Physiol. 2008, 165, 5–18. [Google Scholar] [CrossRef]
- Tucker, S.L.; Talbot, N.J. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 2001, 39, 385–417. [Google Scholar] [CrossRef]
- Heffer, V.; Powelson, M.L.; Johnson, K.B.; Shishkoff, N. Identification of powdery mildew fungi anno 2006. Plant Heath Instr. 2006. [Google Scholar] [CrossRef]
- Sidhu, G.S. Genetics of plant pathogenic fungi. In Advances in Plant Pathology; Ingram, D.S., Williams, P.H., Eds.; Academic Press: Cambridge, MA, USA, 1988; Volume 6. [Google Scholar]
- Gadoury, D.M.; Cadle-Davidson, L.; Wilcox, W.F.; Dry, I.B.; Seem, R.C.; Milgroom, M.G. Grapevine powdery mildew (Erysiphe necator): A fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph: Grapevine powdery mildew. Mol. Plant Pathol. 2012, 13, 1–16. [Google Scholar] [CrossRef]
- Saharan, G.S.; Mehta, N.K.; Meena, P.D. Infection, pathogenesis and disease cycle. In Powdery Mildew Disease of Crucifers: Biology, Ecology and Disease Management; Springer: Singapore, 2019; pp. 95–130. [Google Scholar]
- Jarvis, W.R.; Gubler, W.D.; Grove, G.G. Epidemiology of Powdery Mildews in Agricultural Pathosystems; Bélanger, R.R., Bushnell, W.R., Dik, A.J., Carver, T.L.W., Eds.; APS Press: St. Paul, MN, USA, 2002. [Google Scholar]
- Helfer, S. Rust fungi and global change. New Phytol. 2014, 201, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Aime, M.; Toome, M.; McLaughlin, D. Pucciniomycotina. Systematics and Evolution. In The Mycota; Springer: Berlin/Heidelberg, Germany, 2014; Volume 7A. [Google Scholar]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Lorrain, C.; Gonçalves dos Santos, K.C.; Germain, H.; Hecker, A.; Duplessis, S. Advances in understanding obligate biotrophy in rust fungi. New Phytol. 2019, 222, 1190–1206. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Petre, B.; Frey, P.; Hecker, A.; Rouhier, N.; Duplessis, S. The poplar-poplar rust interaction: Insights from genomics and transcriptomics. J. Pathog. 2011, 2011, 716041. [Google Scholar] [CrossRef]
- Sohn, K.H.; Lei, R.; Nemri, A.; Jones, J.D. The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 2008, 19, 4077–4090. [Google Scholar] [CrossRef]
- Stergiopoulos, I.; de Wit, P.J. Fungal effector proteins. Annu. Rev. Phytopathol. 2009, 47, 233–263. [Google Scholar] [CrossRef]
- Polonio, A.; Pérez-García, A.; Martínez-Cruz, J.; Fernández-Ortuño, D.; de Vicente, A. The haustorium of phytopathogenic fungi: A short overview of a specialized cell of obligate biotrophic plant parasites. In Progress in Botany; Cánovas, F.M., Lüttge, U., Risueño, M.C., Pretzsch, H., Eds.; Springer Nature Switzerland AG: Basel, Switzerland, 2020; Volume 82, ISBN 978-3-030-68619-2. [Google Scholar]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 2010, 22, 3130–3141. [Google Scholar] [CrossRef]
- Pliego, C.; Nowara, D.; Bonciani, G.; Gheorghe, D.M.; Xu, R.; Surana, P.; Whigham, E.; Nettleton, D.; Bogdanove, A.J.; Wise, R.P.; et al. Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol. Plant Microbe Interact. 2013, 26, 633–642. [Google Scholar] [CrossRef]
- Martínez-Cruz, J.; Romero, D.; de la Torre, F.N.; Fernández-Ortuño, D.; Torés, J.A.; de Vicente, A.; Pérez-García, A. The functional characterization of Podosphaera xanthii candidate effector genes reveals novel target functions for fungal pathogenicity. Mol. Plant Microbe Interact. 2018, 31, 914–931. [Google Scholar] [CrossRef]
- Jiang, D.; Zhu, W.; Wang, Y.; Sun, C.; Zhang, K.-Q.; Yang, J. Molecular tools for functional genomics in filamentous fungi: Recent advances and new strategies. Biotech. Adv. 2013, 31, 1562–1574. [Google Scholar] [CrossRef]
- Martínez-Cruz, J.; Romero, D.; Vicente, A.; Pérez-García, A. Transformation of the cucurbit powdery mildew pathogen Podosphaera xanthii by Agrobacterium tumefaciens. New Phytol. 2017, 213, 1961–1973. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Lange, M. VIGS—Genomics goes functional. Trends Plant Sci. 2010, 15, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, M.H.; Keller, Y.; Bouvier, F.; Clary, D.; Camara, B. Functional integration of non-native carotenoids into chloroplasts by viral-derived expression of capsanthin–capsorubin synthase in Nicotiana benthamiana. Plant J. 1998, 14, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.O.; Lim, H.-S.; Bragg, J.; Ganesan, U.; Lee, M.Y. Hordeivirus replication, movement, and pathogenesis. Annu. Rev. Phytopathol. 2009, 47, 385–422. [Google Scholar] [CrossRef]
- Lee, W.-S.; Hammond-Kosack, K.E.; Kanyuka, K. Barley stripe mosaic virus—Mediated tools for investigating gene function in cereal plants and their pathogens: Virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous protein. Plant Physiol. 2012, 160, 582–590. [Google Scholar] [CrossRef]
- Yin, C.; Jurgenson, J.E.; Hulbert, S.H. Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 2011, 24, 554–561. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, J.; Voegele, R.T.; Zhang, J.; Duan, Y.; Luo, H.; Kang, Z. Functional characterization of calcineurin homologs PsCNA1/PsCNB1 in Puccinia striiformis f. sp. tritici using a host-induced RNAi system. PLoS ONE 2012, 7, e49262. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, X.; Yao, J.; Voegele, R.T.; Zhang, Y.; Wang, W.; Huang, L.; Kang, Z. Characterization of protein kinase PsSRPKL, a novel pathogenicity factor in the wheat stripe rust fungus: Kinases and rust pathogenicity. Environ. Microbiol. 2015, 17, 2601–2617. [Google Scholar] [CrossRef]
- Tang, C.; Wei, J.; Han, Q.; Liu, R.; Duan, X.; Fu, Y.; Huang, X.; Wang, X.; Kang, Z. PsANT, the adenine nucleotide translocase of Puccinia striiformis, promotes cell death and fungal growth. Sci. Rep. 2015, 5, 11241. [Google Scholar] [CrossRef]
- Liu, J.; Guan, T.; Zheng, P.; Chen, L.; Yang, Y.; Huai, B.; Li, D.; Chang, Q.; Huang, L.; Kang, Z. An extracellular Zn-Only superoxide dismutase from Puccinia striiformis confers enhanced resistance to host-derived oxidative stress. Environ. Microbiol. 2016, 18, 4118–4135. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Yao, J.; Zhang, Y.; Li, S.; Kang, Z. Characterization of a Ran gene from Puccinia striiformis f. sp. tritici involved in fungal growth and anti-cell death. Sci. Rep. 2016, 6, 35248. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Qi, T.; Yang, Q.; He, F.; Tan, C.; Ma, W.; Voegele, R.T.; Kang, Z.; Guo, J. Host-induced gene silencing of the MAPKK gene PsFUZ7 confers stable resistance to wheat stripe rust. Plant Physiol. 2017, 175, 1853–1863. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, W.; Chu, X.; Sun, Q.; Tan, C.; Yang, Q.; Jiao, M.; Guo, J.; Kang, Z. The transcription factor PstSTE12 is required for virulence of Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 2018, 19, 961–974. [Google Scholar] [CrossRef]
- Qi, T.; Zhu, X.; Tan, C.; Liu, P.; Guo, J.; Kang, Z.; Guo, J. Host-induced gene Silencing of an important pathogenicity factor PsCPK1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnol. J. 2018, 16, 797–807. [Google Scholar] [CrossRef]
- Zhu, X.; Jiao, M.; Guo, J.; Liu, P.; Tan, C.; Yang, Q.; Zhang, Y.; Thomas Voegele, R.; Kang, Z.; Guo, J. A novel MADS-box transcription factor PstMCM1-1 is responsible for full virulence of Puccinia striiformis f. sp. tritici. Environ. Microbiol. 2018, 20, 1452–1463. [Google Scholar] [CrossRef]
- Zhu, X.; Guo, J.; He, F.; Zhang, Y.; Tan, C.; Yang, Q.; Huang, C.; Kang, Z.; Guo, J. Silencing PsKPP4, a MAP kinase kinase kinase gene, reduces pathogenicity of the stripe rust fungus. Mol. Plant Pathol. 2018, 19, 2590–2602. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, J.; Ji, S.; Chen, Z.; Xu, J.; Tang, C.; Chen, S.; Kang, Z.; Wang, X. Candidate effector Pst_8713 impairs the plant immunity and contributes to virulence of Puccinia striiformis f. sp. tritici. Front. Plant Sci. 2018, 9, 1294. [Google Scholar] [CrossRef]
- Qi, T.; Guo, J.; Liu, P.; He, F.; Wan, C.; Islam, M.A.; Tyler, B.M.; Kang, Z.; Guo, J. Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-promoting transcription factor TaLOL2 to defeat ROS-induced defense in wheat. Mol. Plant 2019, 12, 1624–1638. [Google Scholar] [CrossRef]
- Panwar, V.; McCallum, B.; Bakkeren, G. Endogenous silencing of Puccinia triticina pathogenicity genes through in planta-expressed sequences leads to the suppression of rust diseases on wheat. Plant J. 2013, 73, 521–532. [Google Scholar] [CrossRef]
- Yin, C.; Park, J.-J.; Gang, D.R.; Hulbert, S.H. Characterization of a tryptophan 2-monooxygenase gene from Puccinia graminis f. sp. tritici involved in auxin biosynthesis and rust pathogenicity. Mol. Plant Microbe Interact. 2014, 27, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Downey, S.I.; Klages-Mundt, N.L.; Ramachandran, S.; Chen, X.; Szabo, L.J.; Pumphrey, M.; Hulbert, S.H. Identification of promising host-induced silencing targets among genes preferentially transcribed in haustoria of Puccinia. BMC Genomics 2015, 16, 579. [Google Scholar] [CrossRef] [PubMed]
- Margo, R. Genome improvement for rust disease resistance in wheat. In Genome Engineering for Crop Improvement; Sarmah, B.K., Borah, B.K., Eds.; Springer International Publishing: Cham, Switzerland, 2021; ISBN 978-3-030-63371-4. [Google Scholar]
- Govindarajulu, M.; Epstein, L.; Wroblewski, T.; Michelmore, R.W. Host-induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol. J. 2015, 13, 875–883. [Google Scholar] [CrossRef]
- Zhang, W.J.; Pedersen, C.; Kwaaitaal, M.; Gregersen, P.L.; Mørch, S.M.; Hanisch, S.; Kristensen, A.; Fuglsang, A.T.; Collinge, D.B.; Thordal-Christensen, H. Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Mol. Plant Pathol. 2012, 13, 1110–1119. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Pedersen, C.; Schultz-Larsen, T.; Kwaaitaal, M.; Jørgensen, H.J.; Thordal-Christensen, H. The barley powdery mildew candidate secreted effector protein CSEP0105 inhibits the chaperone activity of a small heat shock protein. Plant Physiol. 2015, 168, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Jin, C.; Pei, H.; Zhao, L.; Li, X.; Li, J.; Huang, W.; Fan, R.; Liu, W.; Shen, Q.H. The powdery mildew effector CSEP0027 interacts with barley catalase to regulate host immunity. Front. Plant Sci. 2021, 12, 733237. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, G.B.; Pedersen, C.; Thordal-Christensen, H. Identification of eight effector candidate genes involved in early aggressiveness of the barley powdery mildew fungus. Plant Pathol. 2016, 65, 953–958. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Pedersen, C.; Thordal-Christensen, H. The barley powdery mildew effector candidates CSEP0081 and CSEP0254 promote fungal infection success. PLoS ONE 2016, 11, e157586. [Google Scholar] [CrossRef]
- Li, X.; Jin, C.; Yuan, H.; Huang, W.; Liu, F.; Fan, R.; Xie, J.; Shen, Q.H. The barley powdery mildew effectors CSEP0139 and CSEP0182 suppress cell death and promote B. graminis fungal virulence in plants. Phytopathol. Res. 2021, 3, 7. [Google Scholar] [CrossRef]
- Sanford, J.C. Biolistic plant transformation. Physiol. Plant 1990, 79, 206–209. [Google Scholar] [CrossRef]
- Harwood, W.A. Advances and remaining challenges in the transformation of barley and wheat. J. Exp. Bot. 2012, 63, 1791–1798. [Google Scholar] [CrossRef] [PubMed]
- Gelvin, S.B. Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 2003, 67, 16–37. [Google Scholar] [CrossRef] [PubMed]
- Johansen, L.K.; Carrington, J.C. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 2001, 126, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.G.; Wang, C.H.; Guo, H.S. Application of RNA silencing to plant disease resistance. Silence 2012, 3, 5. [Google Scholar] [CrossRef]
- Bertazzon, N.; Raiola, A.; Castiglioni, C.; Gardiman, M.; Angelini, E.; Borgo, M.; Ferrari, S. Transient silencing of the grapevine gene VvPGIP1 by agroinfiltration with a construct for RNA interference. Plant Cell Rep. 2012, 31, 133–143. [Google Scholar] [CrossRef]
- Martínez-Cruz, J.; Romero, D.; Hierrezuelo, J.; Thon, M.; de Vicente, A.; Pérez-García, A. Effectors with chitinase activity (EWCAs), a family of conserved, secreted fungal chitinases that suppress chitin-triggered immunity. Plant Cell 2021, 33, 1319–1340. [Google Scholar] [CrossRef]
- Polonio, Á.; Fernández-Ortuño, D.; Vicente, A.; Pérez-García, A. A haustorial-expressed lytic polysaccharide monooxygenase from the cucurbit powdery mildew pathogen Podosphaera xanthii contributes to the suppression of chitin-triggered immunity. Mol. Plant. Pathol. 2021, 22, 580–601. [Google Scholar] [CrossRef]
- Martínez-Cruz, J.M.; Polonio, Á.; Zanni, R.; Romero, D.; Gálvez, J.; Fernández-Ortuño, D.; Pérez-García, A. chitin deacetylase, a novel target for the design of agricultural fungicides. J. Fungi 2021, 7, 1009. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V. Exogenous RNAs for gene regulation and plant resistance. Int. J. Mol. Sci. 2019, 20, 2282. [Google Scholar] [CrossRef]
- Dalakouras, A.; Wassenegger, M.; Dadami, E.; Ganopoulos, I.; Pappas, M.L.; Papadopoulou, K. Genetically modified organism-free RNA interference: Exogenous application of RNA molecules in plants. Plant Physiol. 2020, 182, 38–50. [Google Scholar] [CrossRef]
- Mumbanza, F.M.; Kiggundu, A.; Tusiime, G.; Tushemereirwe, W.K.; Niblett, C.; Bailey, A. In vitro antifungal activity of synthetic dsRNA molecules against two pathogens of banana, Fusarium oxysporum f. sp. cubense and Mycosphaerella fijiensis. Pest. Manag. Sci. 2013, 69, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weiberg, A.; Dellota, E.; Yamane, D.; Jin, H. Botrytis small RNA Bc -siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 2017, 14, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Aminedi, R.; Saxena, D.; Gupta, A.; Banerjee, P.; Jain, D.; Chandran, D. Effector mining from the Erysiphe pisi haustorial transcriptome identifies novel candidates involved in pea powdery mildew pathogenesis. Mol. Plant Pathol. 2019, 20, 1506–1522. [Google Scholar] [CrossRef]
- Saito, H.; Sakata, N.; Ishiga, T.; Ishiga, Y. Efficacy of RNA-spray-induced silencing of Phakopsora pachyrhizi chitin synthase genes to control soybean rust. J. Gen. Plant Pathol. 2022, 88, 203–206. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.; Kogel, K.-H.; Jin, H. Cross-kingdom RNA trafficking and environmental RNAi—Nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 2018, 46, 58–64. [Google Scholar] [CrossRef]
- Koch, A.; Kumar, N.; Weber, L.; Keller, H.; Imani, J.; Kogel, K.-H. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase–encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. USA 2013, 110, 19324–19329. [Google Scholar] [CrossRef]
- Cheng, S.; Xie, X.; Xu, Y.; Zhang, C.; Wang, X.; Zhang, J.; Wang, Y. Genetic transformation of a fruit-specific, highly expressed stilbene synthase gene from chinese wild Vitis Quinquangularis. Planta 2016, 243, 1041–1053. [Google Scholar] [CrossRef]
- Guo, X.-Y.; Li, Y.; Fan, J.; Xiong, H.; Xu, F.X.; Shi, J.; Shi, Y.; Zhao, J.Q.; Wang, Y.F.; Cao, X.L.; et al. Host-induced gene silencing of MoAP1 confers broad-spectrum resistance to Magnaporthe oryzae. Front. Plant Sci. 2019, 10, 433. [Google Scholar] [CrossRef]
- Both, M.; Eckert, S.E.; Csukai, M.; Müller, E.; Dimopoulos, G.; Spanu, P.D. Transcript profiles of Blumeria graminis development during infection reveal a cluster of genes that are potential virulence determinants. Mol. Plant Microbe Interact. 2005, 18, 125–133. [Google Scholar] [CrossRef]
- Pedersen, C.; van Themaat, E.V.L.; McGuffin, L.J.; Abbott, J.C.; Burgis, T.A.; Barton, G.; Bindschedler, L.V.; Lu, X.; Maekawa, T.; Wessling, R.; et al. Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 2012, 13, 694. [Google Scholar] [CrossRef]
- Bourras, S.; Praz, C.R.; Spanu, P.D.; Keller, B. Cereal powdery mildew effectors: A complex toolbox for an obligate pathogen. Curr. Opin. Microbiol. 2018, 46, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Fuller, K.K.; Rhodes, J.C. Protein kinase A and fungal virulence: A sinister side to a conserved nutrient sensing pathway. Virulence 2012, 3, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Zhou, X.; Gu, X.; Cao, S.; Wang, C.; Xu, J.-R. The cAMP-PKA pathway regulates growth, sexual and asexual differentiation, and pathogenesis in Fusarium graminearum. Mol. Plant Microbe Intract. 2014, 27, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Song, X.S.; Li, H.P.; Cao, L.H.; Sun, K.; Qiu, X.L.; Xu, Y.B.; Yang, P.; Huang, T.; Zhang, J.B.; et al. Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 2015, 13, 1335–1345. [Google Scholar] [CrossRef]
- Wang, X.; Zhai, T.; Zhang, X.; Tang, C.; Zhuang, R.; Zhao, H.; Xu, Q.; Cheng, Y.; Wang, J.; Duplessis, S.; et al. Two stripe rust effectors impair wheat resistance by suppressing import of host Fe–S protein into chloroplasts. Plant Physiol. 2021, 187, 2530–2543. [Google Scholar] [CrossRef]
- Panwar, V.; Jordan, M.; McCallum, B.; Bakkeren, G. Host-induced silencing of essential genes in Puccinia triticina through transgenic expression of RNAi sequences reduces severity of leaf rust infection in wheat. Plant Biotechnol. J. 2018, 16, 1013–1023. [Google Scholar] [CrossRef]
- Islam, M.T.; Sherif, S.M. RNAi-Based Biofungicides as a promising next-generation strategy for controlling devastating gray mold diseases. Int. J. Mol. Sci. 2020, 21, 2072. [Google Scholar] [CrossRef]
- Qiao, L.; Lan, C.; Capriotti, L.; Ah-Fong, A.; Nino Sanchez, J.; Hamby, R.; Heller, J.; Zhao, H.; Glass, N.L.; Judelson, H.S.; et al. Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 2021, 19, 1756–1768. [Google Scholar] [CrossRef]
- Hu, D.; Chen, Z.; Zhang, C.; Ganiger, M. Reduction of Phakopsora pachyrhizi infection on soybean through host- and spray-induced gene silencing. Mol. Plant Pathol. 2020, 21, 794–807. [Google Scholar] [CrossRef]
- Degnan, R.M.; McTaggart, A.R.; Shuey, L.S.; Pame, L.J.S.; Smith, G.R.; Gardiner, D.M.; Nock, V.; Soffe, R.; Sale, S.; Garrill, A.; et al. Exogenous double-stranded RNA inhibits the infection physiology of rust fungi to reduce symptoms in planta. Mol. Plant Pathol. 2023, 24, 191–207. [Google Scholar] [CrossRef]
- Wang, M.; Jin, H. Spray-induced gene silencing: A powerful innovative strategy for crop protection. Trends Microbiol. 2017, 25, 4–6. [Google Scholar] [CrossRef]
- Taning, C.N.; Arpaia, S.; Christiaens, O.; Dietz-Pfeilstetter, A.; Jones, H.; Mezzetti, B.; Sabbadini, S.; Sorteberg, H.; Sweet, J.; Ventura, V.; et al. RNA-based biocontrol compounds: Current status and perspectives to reach the market. Pest Manag. Sci. 2020, 76, 841–845. [Google Scholar] [CrossRef] [PubMed]
- Landry, M.P.; Mitter, N. How nanocarriers delivering cargos in plants can change the GMO landscape. Nat. Nanotechnol. 2019, 14, 512–514. [Google Scholar] [CrossRef] [PubMed]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.G.; Fletcher, S.J.; Manzie, N.; Robinson, K.E.; Li, P.; Lu, E.; Brosnan, C.A.; Xu, Z.P.; Mitter, N. Foliar application of clay-delivered RNA interference for whitefly control. Nat. Plants 2022, 8, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Mosa, M.A.; Youssef, K. Topical delivery of host induced RNAi silencing by layered double hydroxide nanosheets: An efficient tool to decipher pathogenicity gene function of Fusarium crown and root rot in tomato. Physiol. Mol. Plant Pathol. 2021, 115, 101684. [Google Scholar] [CrossRef]
- Niño-Sánchez, J.; Sambasivam, P.T.; Sawyer, A.; Hamby, R.; Chen, A.; Czislowski, E.; Li, P.; Manzie, N.; Gardiner, D.M.; Ford, R.; et al. BioClayTM prolongs RNA interference-mediated crop protection against Botrytis cinerea. J. Integr. Plant Biol. 2022, 64, 2187–2198. [Google Scholar] [CrossRef]
- Delgado-Martín, J.; Delgado-Olidén, A.; Velasco, L. Carbon dots boost dsRNA delivery in plants and increase local and systemic siRNA production. Int. J. Mol. Sci. 2022, 23, 5338. [Google Scholar] [CrossRef]
- Kostov, K.; Andonova-Lilova, B.; Smagghe, G. Inhibitory activity of carbon quantum dots against Phytophthora infestans and fungal plant pathogens and their effect on dsRNA-induced gene silencing. Biotechnol. Biotechnol. Equip. 2022, 36, 949–959. [Google Scholar] [CrossRef]
- Karny, A.; Zinger, A.; Kajal, A.; Shainsky-Roitman, J.; Schroeder, A. Therapeutic nanoparticles penetrate leaves and deliver nutrients to agricultural crops. Sci. Rep. 2018, 8, 7589. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, H.; Demirer, G.S.; González-Grandío, E.; Fan, C.; Landry, M.P. Engineering DNA nanostructures for siRNA delivery in plants. Nat. Protoc. 2020, 15, 3064–3087. [Google Scholar] [CrossRef] [PubMed]





| Plant Host | Pathogen | Target Gene | Possible Gene Function | Application | Phenotype | References |
|---|---|---|---|---|---|---|
| Hordeum vulgare | Blumeria graminis f. sp. hordei | GTF1 | Cell wall elongation and virulence factor | Virus inoculation by rubbing of barley first leaves | Reduction in haustorium formation | [53] |
| GTF2 | ||||||
| Triticum aestivum | Puccinia striiformis f. sp. tritici | PSTha12J12 | Predicted secreted protein | Virus inoculation by rubbing wheat leaves | Reduction in the expression patternsof the fungal genes | [62] |
| PSTha5A23 | ||||||
| PSTha12H2 | ||||||
| PSTha2A5 | ||||||
| PSTha9F18 | ||||||
| PSTha5A1 | Predicted to code for a chitinase protein | |||||
| PSTha12O3 | Homologous to Uromycesfabae hexose trans-porters | |||||
| PsCNA1 | Calcineurin A-like protein (CNA1) | Slower elongation of fungal hyphae and reduction of the production of uredospore | [63] | |||
| PsCNB1 | Calcineurin B-like protein (CNB1) | |||||
| PsSRPKL | Protein kinase | Reduction of fungal growth and increases of ROS accumulation in host cells | [64] | |||
| PsANT | Adenine nucleotide translocase | Attenuated the growth and development of virulent Pst at the early infection stage | [65] | |||
| PsSOD1 | Zn-only superoxide dismutase | Reduction of the virulence-associated with ROS accumulation | [66] | |||
| PsRan | Small GTP-binding protein | Reduction of the number of haustoria and the length of infection hyphae | [67] | |||
| PsFUZ7 | MAPK kinase | Reduction of initial haustorium formation and elongation of secondary hyphae | [68] | |||
| PstSTE12 | Transcription factor | Reduction in the growth and spread of hyphae in Pst and weakened the virulence of Pst on wheat | [69] | |||
| T. aestivum | P. striiformis f. sp. tritici | PsCPK1 | PKA catalytic subunit | Virus inoculation by rubbing wheat leaves | Reduction in the length of infection hyphae and disease phenotype | [70] |
| PstMCM1-1 | MADX-box transcription factor | Reduction of hyphal extension and haustorium formation | [71] | |||
| PsKPP4 | MAPK kinase | Reduction of haustorium number | [72] | |||
| Pst_8713 | Suppresses host defenses and contributes to the pathogenicity of Pst | Reduction of haustorium number | [73] | |||
| PstGSRE1 | Effector to defeat ROS-associated plant defense by modulating the subcellular compartment of a host immune regulator | Reduction in sporulation and in the fungi biomass | [74] | |||
| Puccinia triticina | PtCYC1 | Cyclophilin | Reduction in fungal growth and disease symptoms | [75] | ||
| PtMAPK1 | MAP kinase | |||||
| PtCNB | Calcineurin regulatory subunit | |||||
| Puccinia graminis f. sp. tritici | Pgt-IaaM | Tryptophan mono-oxygenase | Reduction in fungal growth and in the size of uredinia | [76] | ||
| PGTG_01136 | Predicted glycolytic enzyme | Reduction in fungal growth and in the size of uredinia | [77] | |||
| PGTG_01215 | Probably involved in cellular carbohydrate or sugar metabolism | |||||
| PGTG_03478 | ||||||
| PGTG_14350 | Hypothetical secreted protein with homology to periplasmic components of prokaryotic transport systems | |||||
| PGTG_10731 | Hypothetical proteins | |||||
| PGTG_12890 | ||||||
| PGTG_01304 | Protein involved in thiazole biosynthesis | |||||
| PGTG_16914 | Amino acid permease | |||||
| PGTG_03590 | Secreted protein | |||||
| Pgt-IaaM | Tryptophan 2-monooxygenase enzyme |
| Plant Host | Pathogen | Target Gene | Possible Gene Function | Application | Phenotype | References |
|---|---|---|---|---|---|---|
| H. vulgare | Blumeria graminis f. sp. hordei | Avra10 | Virulence effector | Microprojectile bombardment | Reduction in haustorium formation | [53] |
| BEC1054 | Ribonuclease-like protein | Reduction in haustorium formation | [54] | |||
| BEC1011 | ||||||
| BEC1019 | Metalloprotease | |||||
| BEC1005 | Endo β1-3 glucanase | |||||
| CSEP0055 | Effector involved in secondary penetration events | Reduction in haustorium formation | [80] | |||
| CSEP0105 | Effector proteins | Reduction in haustorium formation | [81] | |||
| CSEP0162 | ||||||
| CSEP0027 | Interacts with barley HvCAT1 to regulate the host immunity to promote fungal virulence | Reduction in haustoria formation | [82] | |||
| CSEP0007 | Possibly involved in penetration and/or establishment of primary haustoria | Reduction in haustoria formation | [83] | |||
| CSEP0025 | ||||||
| CSEP0128 | ||||||
| CSEP0247 | ||||||
| CSEP0345 | ||||||
| CSEP0420 | ||||||
| CSEP0422 | ||||||
| CSEP0081 | Candidate Secreted Effector Proteins | Microprojectile bombardment | Reduction in fungal growth and in haustorium formation | [84] | ||
| CSEP0254 | ||||||
| CSEP0139 | Suppressed cell death triggered by BAX and NtMEK2DD | Reduction in haustoria formation | [85] | |||
| CSEP0182 |
| Plant Host | Pathogen | Target Gene | Possible Gene Function | Application | Phenotype | References |
|---|---|---|---|---|---|---|
| T. aestivum | Puccinia triticina Puccinia graminis and Puccinia striiformis | PtCYC1 | Cyclophilin | Agroinfiltration through the abaxial surface of wheat seedling leaves | Reduction in fungal growth and sporulation | [75] |
| PtMAPK1 | MAP kinase | |||||
| PtCNB | Calcineurin regulatory subunit | |||||
| Cucumis melo | Podosphaera xanthii | PEC007 | Candidate effector | Agroinfiltration of melon cotyledons | Reduction of fungal growth and increasing of the production of hydrogen peroxide by host cells | [55] |
| PEC009 | ||||||
| PEC034 | ||||||
| PEC032 | α-Mannosidase | |||||
| PEC019 | Phospholipid-binding protein | |||||
| PEC054 | Cellulose-binding protein | |||||
| PEC1666 | Chitinase activity | Reduction of fungal growth and increasing of the production of hydrogen peroxide by host cells | [92] | |||
| PEC1961 | ||||||
| PEC2158 | ||||||
| PEC5191 | ||||||
| PHEC27213 | Lytic polysaccharide mono-oxygenase (LPMO) prevents the activation of chitin-triggered immunity | Reduction of fungal growth and increasing production of hydrogen peroxide by host cells | [93] | |||
| PxCDA | chitin deacetylase | Reduction of fungal growth and increasing production of hydrogen peroxide by host cells | [94] |
| Plant Host | Pathogen | Target Gene | Possible Gene Function | Application | Phenotype | References |
|---|---|---|---|---|---|---|
| Pisum sativum | Erysiphe pisi | EpCSEP001 | Virulence factors | Second leaves of pea plants were infiltrated with 100 parts per million (ppm) EpCSEP/CSP-dsRNA | Reduction in disease symptoms | [99] |
| EpCSEP009 | ||||||
| EpCSP083 | ||||||
| C. melo | Podosphaera xanthii | PxCNAP1048 | Presumably involved in glycosylation | Melon cotyledons were infiltrated with dsRNA solutions of the different target genes in concentrations between 100 and 1000 ng ml−1 | Reduction in fungal growth and disease symptoms | [29] |
| PxCNAP10905 | Presumably involved in respiration | |||||
| PxCNAP30520 | ||||||
| PxCNAP8878 | ||||||
| PxCNAP9066 | ||||||
| PxCNAP948 | Presumably involved in efflux transport | |||||
| PxTUB2 | Involved in β-tubulin synthesis | |||||
| PxCYP51 | Involved in ergosterol synthesis | |||||
| Glycine max | Phakopsora pachyrhizi | CHS | Involved in chitin synthases | Soybean plants were infiltrated with 10 ng ml−1 of dsCHS | Reduction in fungal growth and in the number of urediniospores | [100] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padilla-Roji, I.; Ruiz-Jiménez, L.; Bakhat, N.; Vielba-Fernández, A.; Pérez-García, A.; Fernández-Ortuño, D. RNAi Technology: A New Path for the Research and Management of Obligate Biotrophic Phytopathogenic Fungi. Int. J. Mol. Sci. 2023, 24, 9082. https://doi.org/10.3390/ijms24109082
Padilla-Roji I, Ruiz-Jiménez L, Bakhat N, Vielba-Fernández A, Pérez-García A, Fernández-Ortuño D. RNAi Technology: A New Path for the Research and Management of Obligate Biotrophic Phytopathogenic Fungi. International Journal of Molecular Sciences. 2023; 24(10):9082. https://doi.org/10.3390/ijms24109082
Chicago/Turabian StylePadilla-Roji, Isabel, Laura Ruiz-Jiménez, Nisrine Bakhat, Alejandra Vielba-Fernández, Alejandro Pérez-García, and Dolores Fernández-Ortuño. 2023. "RNAi Technology: A New Path for the Research and Management of Obligate Biotrophic Phytopathogenic Fungi" International Journal of Molecular Sciences 24, no. 10: 9082. https://doi.org/10.3390/ijms24109082
APA StylePadilla-Roji, I., Ruiz-Jiménez, L., Bakhat, N., Vielba-Fernández, A., Pérez-García, A., & Fernández-Ortuño, D. (2023). RNAi Technology: A New Path for the Research and Management of Obligate Biotrophic Phytopathogenic Fungi. International Journal of Molecular Sciences, 24(10), 9082. https://doi.org/10.3390/ijms24109082