Proteomics Reveals How the Tardigrade Damage Suppressor Protein Teaches Transfected Human Cells to Survive UV-C Stress
Abstract
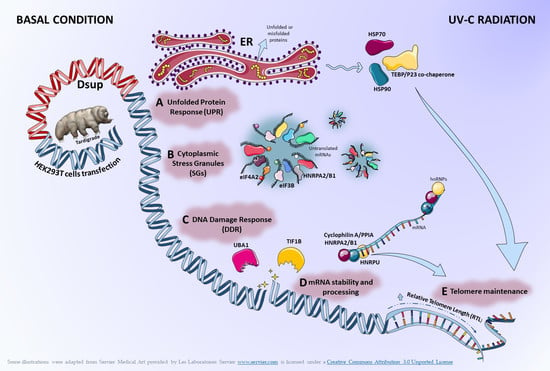
:1. Introduction
2. Results
2.1. In the Absence of External Stimuli, Dsup-Transfected Cells Present a Modulation of Proteins Involved in Protein Folding, Telomere Maintenance, and Metabolic Processes
2.2. Proteins Involved in DNA-Damage Response and in the Regulation of Transcription Are Increased in Dsup+ Cells after 15″ of UV-C Irradiation Followed by 24 h of Recovery
2.3. Targeted Validation of Proteins Belonging to Unfolded Protein Response (UPR), Transcription and Metabolic Regulation, DDR, and Telomere Length Regulation Pathways
3. Discussion
4. Materials and Methods
4.1. Cell Cultures, Dsup Transfection, and UV-C Radiation
4.2. Proteomic Analysis
4.3. Protein Identification by MALDI-ToF Mass Spectrometry
4.4. Telomere Length
4.5. Western Blot
4.6. Statistical and Enrichment Analyses
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hashimoto, T.; Horikawa, D.D.; Saito, Y.; Kuwahara, H.; Kozuka-Hata, H.; Shin-I, T.; Minakuchi, Y.; Ohishi, K.; Motoyama, A.; Aizu, T.; et al. Extremotolerant Tardigrade Genome and Improved Radiotolerance of Human Cultured Cells by Tardigrade-Unique Protein. Nat. Commun. 2016, 7, 12808. [Google Scholar] [CrossRef]
- Degma, P.; Bertolani, R.; Guidetti, R. Actual Checklist of Tardigrada Species; UNIMORE: Modena, Italy, 2019. [Google Scholar]
- Chavez, C.; Cruz-Becerra, G.; Fei, J.; Kassavetis, G.A.; Kadonaga, J.T. The Tardigrade Damage Suppressor Protein Binds to Nucleosomes and Protects DNA from Hydroxyl Radicals. Elife 2019, 8, e47682. [Google Scholar] [CrossRef]
- Hengherr, S.; Worland, M.R.; Reuner, A.; Brümmer, F.; Schill, R.O. High-Temperature Tolerance in Anhydrobiotic Tardigrades Is Limited by Glass Transition. Physiol. Biochem. Zool. 2009, 82, 749–755. [Google Scholar] [CrossRef]
- Ono, F.; Saigusa, M.; Uozumi, T.; Matsushima, Y.; Ikeda, H.; Saini, N.L.; Yamashita, M. Effect of High Hydrostatic Pressure on to Life of the Tiny Animal Tardigrade. J. Phys. Chem. Solids 2008, 69, 2297–2300. [Google Scholar] [CrossRef]
- Arakawa, K. Examples of Extreme Survival: Tardigrade Genomics and Molecular Anhydrobiology. Annu. Rev. Anim. Biosci. 2022, 10, 17–37. [Google Scholar] [CrossRef]
- Kirke, J.; Jin, X.-L.; Zhang, X.-H. Expression of a Tardigrade Dsup Gene Enhances Genome Protection in Plants. Mol. Biotechnol. 2020, 62, 563–571. [Google Scholar] [CrossRef]
- Ricci, C.; Riolo, G.; Marzocchi, C.; Brunetti, J.; Pini, A.; Cantara, S. The Tardigrade Damage Suppressor Protein Modulates Transcription Factor and DNA Repair Genes in Human Cells Treated with Hydroxyl Radicals and UV-C. Biology 2021, 10, 970. [Google Scholar] [CrossRef]
- Abdelzaher, H.; Tawfik, S.M.; Nour, A.; Abdelkader, S.; Elbalkiny, S.T.; Abdelkader, M.; Abbas, W.A.; Abdelnaser, A. Climate Change, Human Health, and the Exposome: Utilizing OMIC Technologies to Navigate an Era of Uncertainty. Front. Public Health 2022, 10, 973000. [Google Scholar] [CrossRef]
- Kciuk, M.; Marciniak, B.; Mojzych, M.; Kontek, R. Focus on UV-Induced DNA Damage and Repair-Disease Relevance and Protective Strategies. Int. J. Mol. Sci. 2020, 21, 7264. [Google Scholar] [CrossRef] [PubMed]
- Gentile, M.; Latonen, L.; Laiho, M. Cell Cycle Arrest and Apoptosis Provoked by UV Radiation-Induced DNA Damage Are Transcriptionally Highly Divergent Responses. Nucleic Acids Res. 2003, 31, 4779–4790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rochette, P.J.; Brash, D.E. Human Telomeres Are Hypersensitive to UV-Induced DNA Damage and Refractory to Repair. PLoS Genet. 2010, 6, e1000926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 Chaperone Machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, T.; Falkowski, M.; Wang, J.J.; Zhang, S.X. Molecular Chaperone ERp29: A Potential Target for Cellular Protection in Retinal and Neurodegenerative Diseases. Adv. Exp. Med. Biol. 2018, 1074, 421–427. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, Y.; Zhang, D. Endoplasmic Reticulum Protein 29 (ERp29) Confers Radioresistance through the DNA Repair Gene, O(6)-Methylguanine DNA-Methyltransferase, in Breast Cancer Cells. Sci. Rep. 2015, 5, 14723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millard, T.H.; Behrendt, B.; Launay, S.; Fütterer, K.; Machesky, L.M. Identification and Characterisation of a Novel Human Isoform of Arp2/3 Complex Subunit P16-ARC/ARPC5. Cell Motil. Cytoskelet. 2003, 54, 81–90. [Google Scholar] [CrossRef]
- Schrank, B.R.; Aparicio, T.; Li, Y.; Chang, W.; Chait, B.T.; Gundersen, G.G.; Gottesman, M.E.; Gautier, J. Nuclear ARP2/3 Drives DNA Break Clustering for Homology-Directed Repair. Nature 2018, 559, 61–66. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, A.; Zheng, L.; Johnathan, A.-F.; Zhang, J.; Zhang, G. The Biological Significance and Regulatory Mechanism of C-Myc Binding Protein 1 (MBP-1). Int. J. Mol. Sci. 2018, 19, 3868. [Google Scholar] [CrossRef] [Green Version]
- Davenport, E.L.; Morgan, G.J.; Davies, F.E. Untangling the Unfolded Protein Response. Cell Cycle 2008, 7, 865–869. [Google Scholar] [CrossRef] [Green Version]
- Qiao, G.; Wu, A.; Chen, X.; Tian, Y.; Lin, X. Enolase 1, a Moonlighting Protein, as a Potential Target for Cancer Treatment. Int. J. Biol. Sci. 2021, 17, 3981–3992. [Google Scholar] [CrossRef]
- Luo, Q.; Jiang, L.; Chen, G.; Feng, Y.; Lv, Q.; Zhang, C.; Qu, S.; Zhu, H.; Zhou, B.; Xiao, X. Constitutive Heat Shock Protein 70 Interacts with α-Enolase and Protects Cardiomyocytes against Oxidative Stress. Free Radic. Res. 2011, 45, 1355–1365. [Google Scholar] [CrossRef]
- Zeng, T.; Cao, Y.; Gu, T.; Chen, L.; Tian, Y.; Li, G.; Shen, J.; Tao, Z.; Lu, L. Alpha-Enolase Protects Hepatocyte Against Heat Stress Through Focal Adhesion Kinase-Mediated Phosphatidylinositol 3-Kinase/Akt Pathway. Front. Genet. 2021, 12, 693780. [Google Scholar] [CrossRef]
- Huang, C.K.; Sun, Y.; Lv, L.; Ping, Y. ENO1 and Cancer. Mol. Ther. Oncolytics 2022, 24, 288–298. [Google Scholar] [CrossRef]
- Didiasova, M.; Schaefer, L.; Wygrecka, M. When Place Matters: Shuttling of Enolase-1 Across Cellular Compartments. Front. Cell Dev. Biol. 2019, 7, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maranto, C.; Perconti, G.; Contino, F.; Rubino, P.; Feo, S.; Giallongo, A. Cellular Stress Induces Cap-Independent Alpha-Enolase/MBP-1 Translation. FEBS Lett. 2015, 589, 2110–2116. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, R.P.; Richa; Kumar, A.; Tyagi, M.B.; Sinha, R.P. Molecular Mechanisms of Ultraviolet Radiation-Induced DNA Damage and Repair. J. Nucleic Acids 2010, 2010, 592980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilenchik, M.M.; Knudson, A.G. Endogenous DNA Double-Strand Breaks: Production, Fidelity of Repair, and Induction of Cancer. Proc. Natl. Acad. Sci. USA 2003, 100, 12871–12876. [Google Scholar] [CrossRef]
- Takahashi, A.; Ohnishi, T. Does GammaH2AX Foci Formation Depend on the Presence of DNA Double Strand Breaks? Cancer Lett. 2005, 229, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA Damage, Repair, and Mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [Green Version]
- López-Camarillo, C.; Ocampo, E.A.; Casamichana, M.L.; Pérez-Plasencia, C.; Alvarez-Sánchez, E.; Marchat, L.A. Protein Kinases and Transcription Factors Activation in Response to UV-Radiation of Skin: Implications for Carcinogenesis. Int. J. Mol. Sci. 2012, 13, 142–172. [Google Scholar] [CrossRef]
- Andrieux, L.O.; Fautrel, A.; Bessard, A.; Guillouzo, A.; Baffet, G.; Langouët, S. GATA-1 Is Essential in EGF-Mediated Induction of Nucleotide Excision Repair Activity and ERCC1 Expression through ERK2 in Human Hepatoma Cells. Cancer Res. 2007, 67, 2114–2123. [Google Scholar] [CrossRef] [Green Version]
- McAvera, R.M.; Crawford, L.J. TIF1 Proteins in Genome Stability and Cancer. Cancers 2020, 12, 2094. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zhang, S.; Gao, X.; Gao, X.; Xu, X.; Lv, Y.; Zhang, Y.; Zhu, Z.; Zhang, C.; Li, Q.; et al. Roles of Kruppel-Associated Box (KRAB)-Associated Co-Repressor KAP1 Ser-473 Phosphorylation in DNA Damage Response. J. Biol. Chem. 2012, 287, 18937–18952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, G.; Kim, J.; Xu, Q.; Leng, Y.; Orkin, S.H.; Elledge, S.J. A Genome-Wide RNAi Screen Identifies a New Transcriptional Module Required for Self-Renewal. Genes Dev. 2009, 23, 837–848. [Google Scholar] [CrossRef] [Green Version]
- Papalazarou, V.; Machesky, L.M. The Cell Pushes Back: The Arp2/3 Complex Is a Key Orchestrator of Cellular Responses to Environmental Forces. Curr. Opin. Cell Biol. 2021, 68, 37–44. [Google Scholar] [CrossRef]
- Byrne, B.M.; Oakley, G.G. Replication Protein A, the Laxative That Keeps DNA Regular: The Importance of RPA Phosphorylation in Maintaining Genome Stability. Semin. Cell Dev. Biol. 2019, 86, 112–120. [Google Scholar] [CrossRef]
- Moudry, P.; Lukas, C.; Macurek, L.; Hanzlikova, H.; Hodny, Z.; Lukas, J.; Bartek, J. Ubiquitin-Activating Enzyme UBA1 Is Required for Cellular Response to DNA Damage. Cell Cycle 2012, 11, 1573–1582. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.-E.; Wani, M.A.; Chen, J.; Zhu, Q.; Wani, G.; El-Mahdy, M.A.; Wani, A.A. Cellular Ubiquitination and Proteasomal Functions Positively Modulate Mammalian Nucleotide Excision Repair. Mol. Carcinog. 2005, 42, 53–64. [Google Scholar] [CrossRef]
- Agarwal, N.; Rinaldetti, S.; Cheikh, B.B.; Zhou, Q.; Hass, E.P.; Jones, R.T.; Joshi, M.; LaBarbera, D.V.; Knott, S.R.V.; Cech, T.R.; et al. TRIM28 Is a Transcriptional Activator of the Mutant TERT Promoter in Human Bladder Cancer. Proc. Natl. Acad. Sci. USA 2021, 118, e2102423118. [Google Scholar] [CrossRef] [PubMed]
- Boltze, C.; Schneider-Stock, R.; Roessner, A.; Quednow, C.; Hoang-Vu, C. Function of HSP90 and P23 in the Telomerase Complex of Thyroid Tumors. Pathol. Res. Pract. 2003, 199, 573–579. [Google Scholar] [CrossRef]
- Forsythe, H.L.; Jarvis, J.L.; Turner, J.W.; Elmore, L.W.; Holt, S.E. Stable Association of Hsp90 and P23, but Not Hsp70, with Active Human Telomerase. J. Biol. Chem. 2001, 276, 15571–15574. [Google Scholar] [CrossRef] [Green Version]
- Fu, D.; Collins, K. Purification of Human Telomerase Complexes Identifies Factors Involved in Telomerase Biogenesis and Telomere Length Regulation. Mol. Cell 2007, 28, 773–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizuno, H.; Honda, M.; Shirasaki, T.; Yamashita, T.; Yamashita, T.; Mizukoshi, E.; Kaneko, S. Heterogeneous Nuclear Ribonucleoprotein A2/B1 in Association with HTERT Is a Potential Biomarker for Hepatocellular Carcinoma. Liver Int. 2012, 32, 1146–1155. [Google Scholar] [CrossRef]
- Xie, X.; Li, M.; Zhou, M.; Chow, S.F.; Tsang, C.K. Pharmacological Preconditioning by TERT Inhibitor BIBR1532 Confers Neuronal Ischemic Tolerance through TERT-Mediated Transcriptional Reprogramming. J. Neurochem. 2021, 159, 690–709. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.-S.; Kim, H.-Y.; Shin, S.-Y.; Kim, Y.-S.; Lee, D.H.; Park, K.M.; Yoon, H.-S. A Cyclophilin A CPR1 Overexpression Enhances Stress Acquisition in Saccharomyces Cerevisiae. Mol. Cells 2010, 29, 567–574. [Google Scholar] [CrossRef]
- Daneri-Becerra, C.; Valeiras, B.; Gallo, L.I.; Lagadari, M.; Galigniana, M.D. Cyclophilin A Is a Mitochondrial Factor That Forms Complexes with P23—Correlative Evidence for an Anti-Apoptotic Action. J. Cell Sci. 2021, 134, jcs253401. [Google Scholar] [CrossRef] [PubMed]
- Lauranzano, E.; Pozzi, S.; Pasetto, L.; Stucchi, R.; Massignan, T.; Paolella, K.; Mombrini, M.; Nardo, G.; Lunetta, C.; Corbo, M.; et al. Peptidylprolyl Isomerase A Governs TARDBP Function and Assembly in Heterogeneous Nuclear Ribonucleoprotein Complexes. Brain 2015, 138, 974–991. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Smith, R. Nuclear Functions of Heterogeneous Nuclear Ribonucleoproteins A/B. Cell. Mol. Life Sci. 2009, 66, 1239–1256. [Google Scholar] [CrossRef]
- Geuens, T.; Bouhy, D.; Timmerman, V. The HnRNP Family: Insights into Their Role in Health and Disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef] [Green Version]
- Haley, B.; Paunesku, T.; Protić, M.; Woloschak, G.E. Response of Heterogeneous Ribonuclear Proteins (HnRNP) to Ionising Radiation and Their Involvement in DNA Damage Repair. Int. J. Radiat. Biol. 2009, 85, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Jia, R. Splicing Factor Poly(RC)-Binding Protein 1 Is a Novel and Distinctive Tumor Suppressor. J. Cell. Physiol. 2018, 234, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Marcelo, A.; Koppenol, R.; de Almeida, L.P.; Matos, C.A.; Nóbrega, C. Stress Granules, RNA-Binding Proteins and Polyglutamine Diseases: Too Much Aggregation? Cell Death Dis. 2021, 12, 592. [Google Scholar] [CrossRef]
- Grosset, C.; Chen, C.Y.; Xu, N.; Sonenberg, N.; Jacquemin-Sablon, H.; Shyu, A.B. A Mechanism for Translationally Coupled MRNA Turnover: Interaction between the Poly(A) Tail and a c-Fos RNA Coding Determinant via a Protein Complex. Cell 2000, 103, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Youn, J.-Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.R.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M.; et al. High-Density Proximity Mapping Reveals the Subcellular Organization of MRNA-Associated Granules and Bodies. Mol. Cell 2018, 69, 517–532. [Google Scholar] [CrossRef]
- Kedersha, N.; Ivanov, P.; Anderson, P. Stress Granules and Cell Signaling: More than Just a Passing Phase? Trends Biochem. Sci. 2013, 38, 494–506. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Jin, X.; Liu, B. The Involvement of Stress Granules in Aging and Aging-Associated Diseases. Aging Cell 2020, 19, e13136. [Google Scholar] [CrossRef] [Green Version]
- Handa, T.; Kundu, D.; Dubey, V.K. Perspectives on Evolutionary and Functional Importance of Intrinsically Disordered Proteins. Int. J. Biol. Macromol. 2023, 224, 243–255. [Google Scholar] [CrossRef]
- Mínguez-Toral, M.; Cuevas-Zuviría, B.; Garrido-Arandia, M.; Pacios, L.F. A Computational Structural Study on the DNA-Protecting Role of the Tardigrade-Unique Dsup Protein. Sci. Rep. 2020, 10, 13424. [Google Scholar] [CrossRef]
- Chavali, S.; Gunnarsson, A.; Babu, M.M. Intrinsically Disordered Proteins Adaptively Reorganize Cellular Matter During Stress. Trends Biochem. Sci. 2017, 42, 410–412. [Google Scholar] [CrossRef]
- Puglia, M.; Landi, C.; Gagliardi, A.; Breslin, L.; Armini, A.; Brunetti, J.; Pini, A.; Bianchi, L.; Bini, L. The Proteome Speciation of an Immortalized Cystic Fibrosis Cell Line: New Perspectives on the Pathophysiology of the Disease. J. Proteom. 2018, 170, 28–42. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Capezzone, M.; Cantara, S.; Marchisotta, S.; Filetti, S.; De Santi, M.M.; Rossi, B.; Ronga, G.; Durante, C.; Pacini, F. Short Telomeres, Telomerase Reverse Transcriptase Gene Amplification, and Increased Telomerase Activity in the Blood of Familial Papillary Thyroid Cancer Patients. J. Clin. Endocrinol. Metab. 2008, 93, 3950–3957. [Google Scholar] [CrossRef] [Green Version]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaba, E.; Landi, C.; Marzocchi, C.; Vantaggiato, L.; Bini, L.; Ricci, C.; Cantara, S. Proteomics Reveals How the Tardigrade Damage Suppressor Protein Teaches Transfected Human Cells to Survive UV-C Stress. Int. J. Mol. Sci. 2023, 24, 11463. https://doi.org/10.3390/ijms241411463
Shaba E, Landi C, Marzocchi C, Vantaggiato L, Bini L, Ricci C, Cantara S. Proteomics Reveals How the Tardigrade Damage Suppressor Protein Teaches Transfected Human Cells to Survive UV-C Stress. International Journal of Molecular Sciences. 2023; 24(14):11463. https://doi.org/10.3390/ijms241411463
Chicago/Turabian StyleShaba, Enxhi, Claudia Landi, Carlotta Marzocchi, Lorenza Vantaggiato, Luca Bini, Claudia Ricci, and Silvia Cantara. 2023. "Proteomics Reveals How the Tardigrade Damage Suppressor Protein Teaches Transfected Human Cells to Survive UV-C Stress" International Journal of Molecular Sciences 24, no. 14: 11463. https://doi.org/10.3390/ijms241411463
APA StyleShaba, E., Landi, C., Marzocchi, C., Vantaggiato, L., Bini, L., Ricci, C., & Cantara, S. (2023). Proteomics Reveals How the Tardigrade Damage Suppressor Protein Teaches Transfected Human Cells to Survive UV-C Stress. International Journal of Molecular Sciences, 24(14), 11463. https://doi.org/10.3390/ijms241411463