A Novel Form of Arginine-Chitosan as Nanoparticles Efficient for siRNA Delivery into Mouse Leukemia Cells
Abstract
:1. Introduction
2. Results
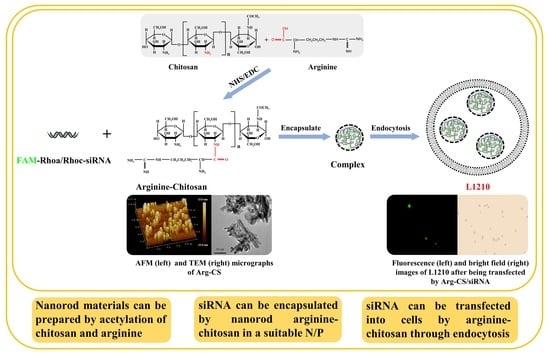
2.1. Modification of Chitosan by Arginine Using EDC/NHS Produced a Kind of Nanoparticle Material
2.2. The Prepared Arginine-Chitosan Nanoparticles Encapsulate siRNA In Vitro via Electrostatic Interaction
2.3. The Arg-CS/Rhoa siRNA Complexes Are Safe for L1210 Cells
2.4. Arginine-Chitosan Nanoparticles Can Deliver siRNA to L1210 Cells
2.5. Rhoa Is Knocked Down in L1210 Cells Transfected by Arg-CS/Rhoa siRNA
3. Discussion
4. Materials and Methods
4.1. Main Reagents
4.2. Synthesis and Characterization of Arginine-Chitosan
4.2.1. Synthesis of Arginine-Chitosan (Arg-CS)
4.2.2. Characterization of Arginine-Chitosan
4.3. Preparation and Characterization of the Arg-CS/siRNA Complex or CS/siRNA Complex
4.4. Cell Culture
4.5. Cell Viability Analysis
4.6. Transfection
4.7. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Subhan, M.A.; Torchilin, V.P. Efficient nanocarriers of siRNA therapeutics for cancer treatment. Transl. Res. 2019, 214, 62–91. [Google Scholar] [CrossRef] [PubMed]
- Cavallaro, G.; Sardo, C.; Craparo, E.F.; Porsio, B.; Giammona, G. Polymeric nanoparticles for siRNA delivery: Production and applications. Int. J. Pharm. 2017, 525, 313–333. [Google Scholar] [CrossRef]
- Yi, L.; Wang, Y.; Lin, G.; Lin, D.; Chen, W.; Huang, Y.; Ye, G. Synthesis of conformation switchable cationic polypeptides based on poly(S-propargyl-cysteine) for use as siRNA delivery. Int. J. Biol. Macromol. 2017, 101, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Kanasty, R.; Dorkin, J.R.; Vegas, A.; Anderson, D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013, 12, 967–977. [Google Scholar] [CrossRef]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Buyens, K.; de Smedt, S.C.; Braeckmans, K.; Demeester, J.; Peeters, L.; van Grunsven, L.A.; de Mollerat du Jeu, X.; Sawant, R.; Torchilin, V.; Farkasova, K.; et al. Liposome based systems for systemic siRNA delivery: Stability in blood sets the requirements for optimal carrier design. J. Control. Release 2012, 158, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef]
- Tai, W.; Gao, X. Functional peptides for siRNA delivery. Adv. Drug Deliv. Rev. 2017, 110–111, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Han, W.; Yuan, Y.; Li, H.; Fu, Z.; Wang, M.; Guan, S.; Wang, L. Design and anti-tumor activity of self-loaded nanocarriers of siRNA. Colloids Surf. B Biointerfaces 2019, 183, 110385. [Google Scholar] [CrossRef]
- Saw, P.E.; Yao, H.; Lin, C.; Tao, W.; Farokhzad, O.C.; Xu, X. Stimuli-Responsive Polymer-Prodrug Hybrid Nanoplatform for Multistage siRNA Delivery and Combination Cancer Therapy. Nano Lett. 2019, 19, 5967–5974. [Google Scholar] [CrossRef]
- Yin, F.; Hu, K.; Chen, Y.; Yu, M.; Wang, D.; Wang, Q.; Yong, K.T.; Lu, F.; Liang, Y.; Li, Z. SiRNA Delivery with PEGylated Graphene Oxide Nanosheets for Combined Photothermal and Genetherapy for Pancreatic Cancer. Theranostics 2017, 7, 1133–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Wu, J.; Liu, Y.; Saw, P.E.; Tao, W.; Yu, M.; Zope, H.; Si, M.; Victorious, A.; Rasmussen, J.; et al. Multifunctional Envelope-Type siRNA Delivery Nanoparticle Platform for Prostate Cancer Therapy. ACS Nano 2017, 11, 2618–2627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabha, B.; Bharadwaj, K.K.; Pati, S.; Choudhury, B.K.; Sarkar, T.; Kari, Z.A.; Edinur, H.A.; Baishya, D.; Atanase, L.I. Development of Polymer-Based Nanoformulations for Glioblastoma Brain Cancer Therapy and Diagnosis: An Update. Polymers 2021, 13, 4114. [Google Scholar] [CrossRef]
- Li, J.; Kataoka, K. Chemo-physical Strategies to Advance the in Vivo Functionality of Targeted Nanomedicine: The Next Generation. J. Am. Chem. Soc. 2021, 143, 538–559. [Google Scholar] [CrossRef] [PubMed]
- Ramishetti, S.; Landesman-Milo, D.; Peer, D. Advances in RNAi therapeutic delivery to leukocytes using lipid nanoparticles. J. Drug Target. 2016, 24, 780–786. [Google Scholar] [CrossRef]
- Kon, E.; Hazan-Halevy, I.; Rosenblum, D.; Cohen, N.; Chatterjee, S.; Veiga, N.; Raanani, P.; Bairey, O.; Benjamini, O.; Nagler, A.; et al. Resveratrol Enhances mRNA and siRNA Lipid Nanoparticles Primary CLL Cell Transfection. Pharmaceutics 2020, 12, 520. [Google Scholar] [CrossRef]
- Antony, R.; Arun, T.; Manickam, S.T.D. A review on applications of chitosan-based Schiff bases. Int. J. Biol. Macromol. 2019, 129, 615–633. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, Q. Recent development of chitosan-based polyelectrolyte complexes with natural polysaccharides for drug delivery. Int. J. Biol. Macromol. 2014, 64, 353–367. [Google Scholar] [CrossRef]
- Van Woensel, M.; Wauthoz, N.; Rosiere, R.; Mathieu, V.; Kiss, R.; Lefranc, F.; Steelant, B.; Dilissen, E.; Van Gool, S.W.; Mathivet, T.; et al. Development of siRNA-loaded chitosan nanoparticles targeting Galectin-1 for the treatment of glioblastoma multiforme via intranasal administration. J. Control. Release 2016, 227, 71–81. [Google Scholar] [CrossRef]
- Gu, J.; Al-Bayati, K.; Ho, E.A. Development of antibody-modified chitosan nanoparticles for the targeted delivery of siRNA across the blood-brain barrier as a strategy for inhibiting HIV replication in astrocytes. Drug Deliv. Transl. Res. 2017, 7, 497–506. [Google Scholar] [CrossRef]
- Aigner, A.; Kogel, D. Nanoparticle/siRNA-based therapy strategies in glioma: Which nanoparticles, which siRNAs? Nanomedicine 2018, 13, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Pitsevicha, G.A.; Malevicha, E.; Kozlovskaya, N.; Doroshenko, I.Y.; Pogorelov, V.E.; Sablinskas, V.; Balevicius, V. Theoretical study of the C-H/O-H stretching vibrations in malonaldehyde. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2015, 145, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Gillis, E.A.L.; Khadijeh, R.; Fridgen, T.D. Structures of hydrated Li+-thymine and Li+-uracil complexes by IRMPD spectroscopy in the N-H/O-H stretching region. J. Phys. Chem. A 2009, 113, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Choe, C.; Lademann, J.; Darvin, M.E. A depth-dependent profile of the lipid conformation and lateral packing order of the stratum corneum in vivo measured using Raman microscopy. Analyst 2016, 141, 1981–1987. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.D.; Qi, S.D.; Lees, R.M.; Xu, L.H. Lamb-dip spectroscopy of the C-N stretching band of methylamine by using frequency-tunable microwave sidebands of CO2 laser lines. Sci. Rep. 2016, 6, 34270. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, L.; Wang, A. Adsorption properties of crosslinked carboxymethyl-chitosan resin with Pb(II) as template ions. J. Hazard. Mater. 2006, 136, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.T.; Bi, Y.; Cao, C. Effects of single bond-ion and single bond-diradical form on the stretching vibration of C=N bridging bond in 4,4′-disubstituted benzylidene anilines. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2016, 163, 96–101. [Google Scholar] [CrossRef]
- Dykxhoorn, D.M.; Lieberman, J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu. Rev. Med. 2005, 56, 401–423. [Google Scholar] [CrossRef] [Green Version]
- Peer, D. A daunting task: Manipulating leukocyte function with RNAi. Immunol. Rev. 2013, 253, 185–197. [Google Scholar] [CrossRef]
- Luo, J.; Li, D.; Wei, D.; Wang, X.; Wang, L.; Zeng, X. RhoA and RhoC are involved in stromal cell-derived factor-1-induced cell migration by regulating F-actin redistribution and assembly. Mol. Cell. Biochem. 2017, 436, 13–21. [Google Scholar] [CrossRef]
- Yuvaraja, G.; Pang, Y.; Chen, D.Y.; Kong, L.J.; Mehmood, S.; Subbaiah, M.V.; Rao, D.S.; Mouli Pavuluri, C.; Wen, J.C.; Reddy, G.M. Modification of chitosan macromolecule and its mechanism for the removal of Pb(II) ions from aqueous environment. Int. J. Biol. Macromol. 2019, 136, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.A.V.; Marangon, C.A.; Martins, V.; Plepis, A.M.G. Chitosan/gelatin films with jatoba resin: Control of properties by vegetal resin inclusion and degree of acetylation modification. Int. J. Biol. Macromol. 2021, 182, 1737–1745. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, M.G.; Soucy, P.A.; Chauhan, R.; Raju, M.V.; Patel, D.N.; Nunn, B.M.; Keynton, M.A.; Ehringer, W.D.; Nantz, M.H.; Keynton, R.S.; et al. Release-Modulated Antioxidant Activity of a Composite Curcumin-Chitosan Polymer. Biomacromolecules 2016, 17, 1253–1260. [Google Scholar] [CrossRef]
- Aleem, A.R.; Shahzadi, L.; Tehseen, S.; Alvi, F.; Chaudhry, A.A.; Rehman, I.U.; Yar, M. Amino acids loaded chitosan/collagen based new membranes stimulate angiogenesis in chorioallantoic membrane assay. Int. J. Biol. Macromol. 2019, 140, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Zhu, D.; Liu, L.; Dong, X.; Zhang, H.; Leng, X. Evaluation of the coagulation properties of arginine-chitosan/DNA nanoparticles. J. Biomed. Mater. Res. B Appl. Biomater. 2010, 95, 374–379. [Google Scholar] [CrossRef]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.G.; Yao, K.D. Chitosan and its derivatives—A promising non-viral vector for gene transfection. J. Control. Release 2002, 83, 1–11. [Google Scholar] [CrossRef]
- Serrano-Sevilla, I.; Artiga, A.; Mitchell, S.G.; de Matteis, L.; de la Fuente, J.M. Natural Polysaccharides for siRNA Delivery: Nanocarriers Based on Chitosan, Hyaluronic Acid, and Their Derivatives. Molecules 2019, 24, 2570. [Google Scholar] [CrossRef] [Green Version]
- Lungwitz, U.; Breunig, M.; Blunk, T.; Gopferich, A. Polyethylenimine-based non-viral gene delivery systems. Eur. J. Pharm. Biopharm. 2005, 60, 247–266. [Google Scholar] [CrossRef]
- Labala, S.; Jose, A.; Venuganti, V.V. Transcutaneous iontophoretic delivery of STAT3 siRNA using layer-by-layer chitosan coated gold nanoparticles to treat melanoma. Colloids Surf. B Biointerfaces 2016, 146, 188–197. [Google Scholar] [CrossRef]
- Zhu, Y.; Yaylayan, V.A. Interaction of free arginine and guanidine with glucose under thermal processing conditions and formation of Amadori-derived imidazolones. Food Chem. 2017, 220, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Hu, Q.; Kondoh, A.; Terada, M. Enantioselective Protonation: Hydrophosphinylation of 1,1-Vinyl Azaheterocycle N-Oxides Catalyzed by Chiral Bis(guanidino)iminophosphorane Organosuperbase. Angew. Chem. Int. Ed. Engl. 2021, 60, 1417–1422. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xu, Z.; Chen, S.; Gu, W.; Chen, L.; Li, Y. Arginine-chitosan/DNA self-assemble nanoparticles for gene delivery: In vitro characteristics and transfection efficiency. Int. J. Pharm. 2008, 359, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Amornwittawat, N.; Juwita, V.; Kao, Y.; Duman, J.G.; Pascal, T.A.; Goddard, W.A.; Wen, X. Arginine, a key residue for the enhancing ability of an antifreeze protein of the beetle Dendroides canadensis. Biochemistry 2009, 48, 9696–9703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, M.; Rusnati, M.; Presta, M.; Giacca, M. Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J. Biol. Chem. 2001, 276, 3254–3261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Shi, H.; Zhu, P.; Wei, Y.; Hao, J. Ammonium Polyphosphate with High Specific Surface Area by Assembling Zeolite Imidazole Framework in EVA Resin: Significant Mechanical Properties, Migration Resistance, and Flame Retardancy. Polymers 2020, 12, 534. [Google Scholar] [CrossRef] [Green Version]
- Garcia, B.B.M.; Mertins, O.; Silva, E.R.D.; Mathews, P.D.; Han, S.W. Arginine-modified chitosan complexed with liposome systems for plasmid DNA delivery. Colloids Surf. B Biointerfaces 2020, 193, 111131. [Google Scholar] [CrossRef]






| Sample Name | Mass Ratio (m/m) | DSL (nm) | PDI | Zeta Potential (mV) |
|---|---|---|---|---|
| siRNA | — | 450.87 ± 53.67 | 0.34 ± 0.17 | −1.67 ± 0.17 |
| CS/siRNA | 1:0 | 483.13 ± 82.34 | 0.33 ± 0.31 | 30.95 ± 2.33 |
| 1:1 | 273.17 ± 26.32 | 0.62 ± 0.11 | 1.74 ± 0.95 | |
| 10:1 | 1299.13 ± 450.22 | 0.73 ± 0.21 | 17.67 ± 2.66 | |
| 20:1 | 1673.13 ± 261.43 | 1.10 ± 0.36 | 17.01 ± 3.93 | |
| 50:1 | 1865.80 ± 99.50 | 1.75 ± 0.18 | 21.5 ± 0.95 | |
| 100:1 | 1122.23 ± 277.23 | 0.66 ± 0.32 | 20.01 ± 6.05 | |
| Arg-CS/siRNA | 1:0 | 75.76 ± 12.07 | 0.43 ± 0.11 | 23.24 ± 1.91 |
| 1:1 | 124.83 ± 11.58 | 0.60 ± 0.39 | 0.57 ± 0.14 | |
| 10:1 | 453.87 ± 100.75 | 0.63 ± 0.12 | 2.23 ± 0.08 | |
| 20:1 | 440.30 ± 178.21 | 0.73 ± 0.07 | 8.62 ± 0.53 | |
| 50:1 | 1261.90 ± 438.88 | 0.68 ± 0.36 | 9.2 ± 0.64 | |
| 100:1 | 296.83 ± 5.13 | 0.65 ± 0.11 | 16.63 ± 1.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, J.; Chen, J.; Liu, Y.; He, Y.; Dong, W. A Novel Form of Arginine-Chitosan as Nanoparticles Efficient for siRNA Delivery into Mouse Leukemia Cells. Int. J. Mol. Sci. 2023, 24, 1040. https://doi.org/10.3390/ijms24021040
Luo J, Chen J, Liu Y, He Y, Dong W. A Novel Form of Arginine-Chitosan as Nanoparticles Efficient for siRNA Delivery into Mouse Leukemia Cells. International Journal of Molecular Sciences. 2023; 24(2):1040. https://doi.org/10.3390/ijms24021040
Chicago/Turabian StyleLuo, Jixian, Jiangfeng Chen, Yan Liu, Yongji He, and Wenjuan Dong. 2023. "A Novel Form of Arginine-Chitosan as Nanoparticles Efficient for siRNA Delivery into Mouse Leukemia Cells" International Journal of Molecular Sciences 24, no. 2: 1040. https://doi.org/10.3390/ijms24021040
APA StyleLuo, J., Chen, J., Liu, Y., He, Y., & Dong, W. (2023). A Novel Form of Arginine-Chitosan as Nanoparticles Efficient for siRNA Delivery into Mouse Leukemia Cells. International Journal of Molecular Sciences, 24(2), 1040. https://doi.org/10.3390/ijms24021040