Lunaemycins, New Cyclic Hexapeptide Antibiotics from the Cave Moonmilk-Dweller Streptomyces lunaelactis MM109T
Abstract
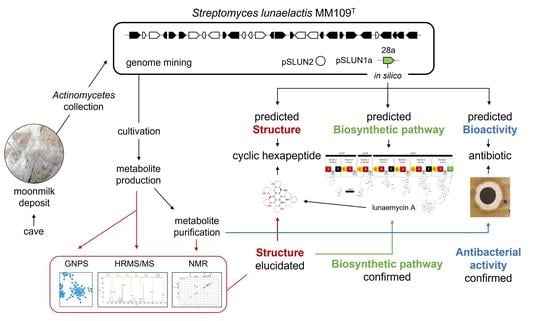
:1. Introduction
2. Results and Discussion
2.1. Sequence Analysis of BGC 28a and Predicted Structure of Its Associated Natural Product
2.2. Structure Elucidation of Lunaemycins
2.2.1. MS/MS-Based Networking and Peptidogenomics Guided Genome Mining
2.2.2. Structure Elucidation by NMR Spectroscopy
2.3. Structural Diversity of Lunaemycins
2.4. Antibacterial Activity of Lunaemycins
3. Materials and Methods
3.1. Bacterial Strains and Culture Conditions
3.2. Bioinformatics and In Silico Analyzes
3.3. Compound Identification by Ultra-Performance Liquid Chromatography–Tandem Mass Spectrometry (UPLC–MS/MS)
3.4. Nuclear Magnetic Resonance (NMR)
3.5. Antibacterial Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. LC Method
Appendix B. HPLC Method
Appendix C. SP-HPLC Method
Appendix D. UPLC Method
Appendix E. MS Method
References
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2015, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Hopwood, D.A. Streptomyces in Nature and Medicine: The Antibiotic Makers; Oxford University Press: Oxford, NY, USA, 2007; ISBN 978-0-19-515066-7. [Google Scholar]
- van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; van Wezel, G.P. Chemical Ecology of Antibiotic Production by Actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, M.L.-Y.; Tan, L.T.-H.; Letchumanan, V.; He, Y.-W.; Fang, C.-M.; Chan, K.-G.; Law, J.W.-F.; Lee, L.-H. The Extremophilic Actinobacteria: From Microbes to Medicine. Antibiot. Basel Switz. 2021, 10, 682. [Google Scholar] [CrossRef] [PubMed]
- Axenov-Gibanov, D.V.; Voytsekhovskaya, I.V.; Tokovenko, B.T.; Protasov, E.S.; Gamaiunov, S.V.; Rebets, Y.V.; Luzhetskyy, A.N.; Timofeyev, M.A. Actinobacteria Isolated from an Underground Lake and Moonmilk Speleothem from the Biggest Conglomeratic Karstic Cave in Siberia as Sources of Novel Biologically Active Compounds. PLoS ONE 2016, 11, e0149216. [Google Scholar] [CrossRef]
- Maciejewska, M.; Całusińska, M.; Cornet, L.; Adam, D.; Pessi, I.S.; Malchair, S.; Delfosse, P.; Baurain, D.; Barton, H.A.; Carnol, M.; et al. High-Throughput Sequencing Analysis of the Actinobacterial Spatial Diversity in Moonmilk Deposits. Antibiotics 2018, 7, E27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Cho, Y.-J.; Jung, D.; Jo, K.; Lee, J.-S. Microbial Diversity of Baeg-Nyong Cave and Characterization of the Antibiotics Extracted from Streptomyces Exfoliatus. FASEB J. 2019, 33, 637.4. [Google Scholar] [CrossRef]
- Rangseekaew, P.; Pathom-Aree, W. Cave Actinobacteria as Producers of Bioactive Metabolites. Front. Microbiol. 2019, 10, 387. [Google Scholar] [CrossRef] [Green Version]
- Adam, D.; Maciejewska, M.; Naômé, A.; Martinet, L.; Coppieters, W.; Karim, L.; Baurain, D.; Rigali, S. Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits. Antibiotics 2018, 7, E28. [Google Scholar] [CrossRef] [Green Version]
- Maciejewska, M.; Adam, D.; Martinet, L.; Naômé, A.; Całusińska, M.; Delfosse, P.; Carnol, M.; Barton, H.A.; Hayette, M.-P.; Smargiasso, N.; et al. A Phenotypic and Genotypic Analysis of the Antimicrobial Potential of Cultivable Streptomyces Isolated from Cave Moonmilk Deposits. Front. Microbiol. 2016, 7, 1455. [Google Scholar] [CrossRef] [Green Version]
- Borsato, A.; Frisia, S.; Jones, B.; Van Der Borg, K. Calcite Moonmilk: Crystal Morphology and Environment of Formation in Caves in the Italian Alps. J. Sediment. Res. 2000, 70, 1171–1182. [Google Scholar] [CrossRef]
- Cañaveras, J.C.; Cuezva, S.; Sanchez-Moral, S.; Lario, J.; Laiz, L.; Gonzalez, J.M.; Saiz-Jimenez, C. On the Origin of Fiber Calcite Crystals in Moonmilk Deposits. Naturwissenschaften 2006, 93, 27–32. [Google Scholar] [CrossRef]
- Reinbacher, W.R. Is It Gnome, Is It Berg, Is It Mont, It It Mond? An Updated View of the Origin and Etymology of Moonmilk. Bull. Natl. Speleol. Soc. 1994, 56, 1–13. [Google Scholar]
- Miller, A.Z.; Dionísio, A.; Jurado, V.; Cuezva, S.; Sanchez-Moral, S.; Cañaveras, J.C.; Saiz-Jimenez, C. Biomineralization by cave dwelling microorganisms. In Advances in Geochemistry Research; Sanjurjo Sanchéz, J., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2013; Volume 5, pp. 77–105. [Google Scholar]
- Maciejewska, M.; Adam, D.; Naômé, A.; Martinet, L.; Tenconi, E.; Całusińska, M.; Delfosse, P.; Hanikenne, M.; Baurain, D.; Compère, P.; et al. Assessment of the Potential Role of Streptomyces in Cave Moonmilk Formation. Front. Microbiol. 2017, 8, 1181. [Google Scholar] [CrossRef] [Green Version]
- Cirigliano, A.; Tomassetti, M.C.; Di Pietro, M.; Mura, F.; Maneschi, M.L.; Gentili, M.D.; Cardazzo, B.; Arrighi, C.; Mazzoni, C.; Negri, R.; et al. Calcite Moonmilk of Microbial Origin in the Etruscan Tomba Degli Scudi in Tarquinia, Italy. Sci. Rep. 2018, 8, 15839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifan, M.; Berenjian, A. Microbially Induced Calcium Carbonate Precipitation: A Widespread Phenomenon in the Biological World. Appl. Microbiol. Biotechnol. 2019, 103, 4693–4708. [Google Scholar] [CrossRef] [PubMed]
- Portillo, M.C.; Gonzalez, J.M. Moonmilk Deposits Originate from Specific Bacterial Communities in Altamira Cave (Spain). Microb. Ecol. 2011, 61, 182–189. [Google Scholar] [CrossRef] [Green Version]
- Spötl, C. Moonmilk as a Human and Veterinary Medicine: Evidence of Past Artisan Mining in Caves of the Austrian Alps. Int. J. Speleol. 2018, 47, 127–135. [Google Scholar] [CrossRef] [Green Version]
- Maciejewska, M.; Pessi, I.S.; Arguelles-Arias, A.; Noirfalise, P.; Luis, G.; Ongena, M.; Barton, H.; Carnol, M.; Rigali, S. Streptomyces Lunaelactis Sp. Nov., a Novel Ferroverdin A-Producing Streptomyces Species Isolated from a Moonmilk Speleothem. Antonie Van Leeuwenhoek 2015, 107, 519–531. [Google Scholar] [CrossRef]
- Naômé, A.; Maciejewska, M.; Calusinska, M.; Martinet, L.; Anderssen, S.; Adam, D.; Tenconi, E.; Deflandre, B.; Coppieters, W.; Karim, L.; et al. Complete Genome Sequence of Streptomyces Lunaelactis MM109T, Isolated from Cave Moonmilk Deposits. Genome. Announc. 2018, 6, e00435-18. [Google Scholar] [CrossRef] [Green Version]
- Martinet, L.; Naômé, A.; Baiwir, D.; De Pauw, E.; Mazzucchelli, G.; Rigali, S. On the Risks of Phylogeny-Based Strain Prioritization for Drug Discovery: Streptomyces Lunaelactis as a Case Study. Biomolecules 2020, 10, E1027. [Google Scholar] [CrossRef] [PubMed]
- Martinet, L.; Baiwir, D.; Mazzucchelli, G.; Rigali, S. Structure of New Ferroverdins Recruiting Unconventional Ferrous Iron Chelating Agents. Biomolecules 2022, 12, 752. [Google Scholar] [CrossRef] [PubMed]
- Martinet, L.; Naômé, A.; Deflandre, B.; Maciejewska, M.; Tellatin, D.; Tenconi, E.; Smargiasso, N.; de Pauw, E.; van Wezel, G.P.; Rigali, S. A Single Biosynthetic Gene Cluster Is Responsible for the Production of Bagremycin Antibiotics and Ferroverdin Iron Chelators. mBio 2019, 10, e01230-19. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Wang, Z.; Huang, H.; Luo, M.; Zuo, D.; Wang, B.; Sun, A.; Cheng, Y.-Q.; Zhang, C.; Ju, J. Biosynthesis of Himastatin: Assembly Line and Characterization of Three Cytochrome P450 Enzymes Involved in the Post-Tailoring Oxidative Steps. Angew. Chem. Int. Ed. Engl. 2011, 50, 7797–7802. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Li, Q.; Qin, X.; Ju, J.; Ma, J. Enhancement of Himastatin Bioproduction via Inactivation of Atypical Repressors in Streptomyces Hygroscopicus. Metab. Eng. Commun. 2019, 8, e00084. [Google Scholar] [CrossRef] [PubMed]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Navarro-Muñoz, J.C.; Terlouw, B.R.; van der Hooft, J.J.J.; van Santen, J.A.; Tracanna, V.; Suarez Duran, H.G.; Pascal Andreu, V.; et al. MIBiG 2.0: A Repository for Biosynthetic Gene Clusters of Known Function. Nucleic Acids Res. 2020, 48, D454–D458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimori, D.G.; Hrvatin, S.; Neumann, C.S.; Strieker, M.; Marahiel, M.A.; Walsh, C.T. Cloning and Characterization of the Biosynthetic Gene Cluster for Kutznerides. Proc. Natl. Acad. Sci. USA 2007, 104, 16498–16503. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.-T.; Lamsa, A.; Wong, W.R.; Boudreau, P.D.; Kersten, R.; Peng, Y.; Moree, W.J.; Duggan, B.M.; Moore, B.S.; Gerwick, W.H.; et al. MS/MS-Based Networking and Peptidogenomics Guided Genome Mining Revealed the Stenothricin Gene Cluster in Streptomyces Roseosporus. J. Antibiot. 2014, 67, 99–104. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Wang, L.; Wan, D.; Qi, J.; Gong, R.; Deng, Z.; Chen, W. Characterization of the Aurantimycin Biosynthetic Gene Cluster and Enhancing Its Production by Manipulating Two Pathway-Specific Activators in Streptomyces Aurantiacus JA 4570. Microb. Cell Factories 2016, 15, 160. [Google Scholar] [CrossRef] [Green Version]
- Broberg, A.; Menkis, A.; Vasiliauskas, R. Kutznerides 1−4, Depsipeptides from the Actinomycete Kutzneria Sp. 744 Inhabiting Mycorrhizal Roots of Picea Abies Seedlings. J. Nat. Prod. 2006, 69, 97–102. [Google Scholar] [CrossRef]
- Gräfe, U.; Schlegel, R.; Ritzau, M.; Ihn, W.; Dornberger, K.; Stengel, C.; Fleck, W.F.; Gutsche, W.; Härtl, A.; Paulus, E.F. Aurantimycins, New Depsipeptide Antibiotics from Streptomyces Aumntiacus IMET 43917 Production, Isolation, Structure Elucidation, and Biological Activity. J. Antibiot. 1995, 48, 119–125. [Google Scholar] [CrossRef] [Green Version]
- Leet, J.E.; Schroeder, D.R.; Golik, J.; Matson, J.A.; Doyle, T.W.; Lam, K.S.; Hill, S.E.; Lee, M.S.; Whitney, J.L.; Krishnan, B.S. Himastatin, a New Antitumor Antibiotic from Streptomyces Hygroscopicus. III. Structural Elucidation. J. Antibiot. 1996, 49, 299–311. [Google Scholar] [CrossRef]
- Hasenböhler, A.; Kneifel, H.; König, W.A.; Zähner, H.; Zeiler, H.J. [Metabolic products of microorganisms. 134. Stenothricin, a new inhibitor of the bacterial cell wall synthesis (author’s transl)]. Arch. Microbiol. 1974, 99, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Neumann, C.S.; Jiang, W.; Heemstra, J.R., Jr.; Gontang, E.A.; Kolter, R.; Walsh, C.T. Biosynthesis of Piperazic Acid via N5-Hydroxy-Ornithine in Kutzneria Spp. 744. ChemBioChem 2012, 13, 972–976. [Google Scholar] [CrossRef]
- Du, Y.-L.; He, H.-Y.; Higgins, M.A.; Ryan, K.S. A Heme-Dependent Enzyme Forms the Nitrogen–Nitrogen Bond in Piperazate. Nat. Chem. Biol. 2017, 13, 836–838. [Google Scholar] [CrossRef]
- Morgan, K.D.; Andersen, R.J.; Ryan, K.S. Piperazic Acid-Containing Natural Products: Structures and Biosynthesis. Nat. Prod. Rep. 2019, 36, 1628–1653. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.-W.; Niikura, H.; Morgan, K.D.; Vacariu, C.M.; Andersen, R.J.; Ryan, K.S. Free Piperazic Acid as a Precursor to Nonribosomal Peptides. J. Am. Chem. Soc. 2022, 144, 13556–13564. [Google Scholar] [CrossRef] [PubMed]
- Chevrette, M.G.; Aicheler, F.; Kohlbacher, O.; Currie, C.R.; Medema, M.H. SANDPUMA: Ensemble Predictions of Nonribosomal Peptide Chemistry Reveal Biosynthetic Diversity across Actinobacteria. Bioinformatics 2017, 33, 3202–3210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Wang, Y.; Huang, T.; Tao, M.; Deng, Z.; Lin, S. Identification and Characterization of the Biosynthetic Gene Cluster of Polyoxypeptin A, a Potent Apoptosis Inducer. BMC Microbiol. 2014, 14, 30. [Google Scholar] [CrossRef] [Green Version]
- Pohlmann, V.; Marahiel, M.A. δ-Amino Group Hydroxylation of L-Ornithine during Coelichelin Biosynthesis. Org. Biomol. Chem. 2008, 6, 1843–1848. [Google Scholar] [CrossRef]
- Wei, H.; Lin, Z.; Li, D.; Gu, Q.; Zhu, T. [OSMAC (one strain many compounds) approach in the research of microbial metabolites—A review]. Wei Sheng Wu Xue Bao 2010, 50, 701–709. [Google Scholar]
- Rigali, S.; Anderssen, S.; Naômé, A.; van Wezel, G.P. Cracking the Regulatory Code of Biosynthetic Gene Clusters as a Strategy for Natural Product Discovery. Biochem. Pharmacol. 2018, 153, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Rigali, S.; Titgemeyer, F.; Barends, S.; Mulder, S.; Thomae, A.W.; Hopwood, D.A.; van Wezel, G.P. Feast or Famine: The Global Regulator DasR Links Nutrient Stress to Antibiotic Production by Streptomyces. EMBO Rep. 2008, 9, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Świątek, M.A.; Urem, M.; Tenconi, E.; Rigali, S.; van Wezel, G.P. Engineering of N-Acetylglucosamine Metabolism for Improved Antibiotic Production in Streptomyces Coelicolor A3(2) and an Unsuspected Role of NagA in Glucosamine Metabolism. Bioengineered 2012, 3, 280–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, N.; Morimoto, K.; Naganawa, H.; Takita, T.; Hamada, M.; Maeda, K.; Takeuchi, T.; Umezawa, H. ANTRIMYCIN, A NEW PEPTIDE ANTIBIOTIC. J. Antibiot. (Tokyo) 1981, 34, 1613–1614. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Huang, P.; Ren, B.; Song, Z.; Zhu, G.; He, W.; Zhang, J.; Oyeleye, A.; Dai, H.; Zhang, L.; et al. Antibacterial Polyene-Polyol Macrolides and Cyclic Peptides from the Marine-Derived Streptomyces Sp. MS110128. Appl. Microbiol. Biotechnol. 2021, 105, 4975–4986. [Google Scholar] [CrossRef]
- Moumbock, A.F.A.; Gao, M.; Qaseem, A.; Li, J.; Kirchner, P.A.; Ndingkokhar, B.; Bekono, B.D.; Simoben, C.V.; Babiaka, S.B.; Malange, Y.I.; et al. StreptomeDB 3.0: An Updated Compendium of Streptomycetes Natural Products. Nucleic. Acids Res. 2021, 49, D600–D604. [Google Scholar] [CrossRef]
- van Santen, J.A.; Poynton, E.F.; Iskakova, D.; McMann, E.; Alsup, T.A.; Clark, T.N.; Fergusson, C.H.; Fewer, D.P.; Hughes, A.H.; McCadden, C.A.; et al. The Natural Products Atlas 2.0: A Database of Microbially-Derived Natural Products. Nucleic Acids Res. 2022, 50, D1317–D1323. [Google Scholar] [CrossRef]
- Peoples, A.S.; Ling, L.L.; Lewis, K.; Zhang, Z. Novel Antibiotics Patent US-2011136752-A1 PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/patent/US-2011136752-A1 (accessed on 31 October 2022).
- Tenconi, E.; Rigali, S. Self-Resistance Mechanisms to DNA-Damaging Antitumor Antibiotics in Actinobacteria. Curr. Opin. Microbiol. 2018, 45, 100–108. [Google Scholar] [CrossRef]
- Besier, S.; Ludwig, A.; Zander, J.; Brade, V.; Wichelhaus, T.A. Linezolid Resistance in Staphylococcus Aureus: Gene Dosage Effect, Stability, Fitness Costs, and Cross-Resistances. Antimicrob. Agents Chemother. 2008, 52, 1570–1572. [Google Scholar] [CrossRef] [Green Version]
- Tedim, A.P.; Lanza, V.F.; Rodríguez, C.M.; Freitas, A.R.; Novais, C.; Peixe, L.; Baquero, F.; Coque, T.M. Fitness Cost of Vancomycin-Resistant Enterococcus Faecium Plasmids Associated with Hospital Infection Outbreaks. J. Antimicrob. Chemother. 2021, 76, 2757–2764. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Röttig, M.; Medema, M.H.; Blin, K.; Weber, T.; Rausch, C.; Kohlbacher, O. NRPSpredictor2--a Web Server for Predicting NRPS Adenylation Domain Specificity. Nucleic Acids Res. 2011, 39, W362–W367. [Google Scholar] [CrossRef] [PubMed]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K. CLSI Methods Development and Standardization Working Group Best Practices for Evaluation of Antimicrobial Susceptibility Tests PMC. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5869819/ (accessed on 27 November 2022).
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Dieases (ESCMID). EUCAST Definitive Document E.Def 1.2, May 2000: Terminology Relating to Methods for the Determination of Susceptibility of Bacteria to Antimicrobial Agents. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2000, 6, 503–508. [Google Scholar] [CrossRef] [Green Version]
- Gubbens, J.; Wu, C.; Zhu, H.; Filippov, D.V.; Florea, B.I.; Rigali, S.; Overkleeft, H.S.; van Wezel, G.P. Intertwined Precursor Supply during Biosynthesis of the Catecholate-Hydroxamate Siderophores Qinichelins in Streptomyces Sp. MBT76. ACS Chem. Biol. 2017, 12, 2756–2766. [Google Scholar] [CrossRef] [PubMed]









| ORFs | Closest Homologue in MiBIG Database (Protein ID) | Homology (Id/Cov) | Proposed Function | |
|---|---|---|---|---|
| Locus Tag Accession No. | Name Size (aa) | |||
| SLUN_RS38410 WP_108155188.1 | Lun1 177 | TmcL, hypothetical protein, α,β-epoxyketone BGC of Streptomyces chromofuscus (CUX96959.1) | 64/100 | Unknown |
| SLUN_RS38415 WP_108155189.1 | Lun2 278 | DUF4097 family beta strand repeat-containing protein, α,β-epoxyketone BGC of Streptomyces chromofuscus (CUX96960.1) | 48/100 | Unknown |
| SLUN_RS38420 WP_108155190.1 | Lun3 416 | Cytochrome P450, streptovaricin BGC of Streptomyces spectabilis (ASZ00144.1) | 61/97 | Product modification |
| SLUN_RS38425 WP_108155191.1 | Lun4 76 | Ferredoxin, caniferolide A BGC from Streptomyces caniferus (QBF51761.1) | 52/82 | Product modification |
| SLUN_RS38430 WP_254708284.1 | Lun5 179 | GtrA family protein, monensin BGC from Streptomyces cinnamonensis (AAO65788.1) | 60/82 | Unknown |
| SLUN_RS38435 WP_159100457.1 | Lun6 746 | MPPL family transporter, alnumycin A BGC from Streptomyces sp. CM020 (ACI88887.1) | 58/98 | Exporter |
| SLUN_RS38440 WP_108155192.1 | Lun7 409 | MfnN, cytochrome P450, marformycin A BGC from Streptomyces drozdowiczii (AJV88386.1) | 37/100 | product modification |
| SLUN_RS38445 WP_108155193.1 | Lun8 157 | Hypothetical protein, no hit in MiBIG, closest to Streptomyces sp. Ag82_O1-15 (WP_095849664.1) | 47/88 | Unknown |
| SLUN_RS38450 WP_108155194.1 | Lun9 531 | MFS transporter, siamycin I BGC from Streptomyces sp. (BBD82029.1) | 72/93 | Resistance export |
| SLUN_RS38455 WP_108155195.1 | Lun10 241 | TetR/AcrR family transcriptional regulator, siamycin I BGC from Streptomyces sp. (BBD82030.1) | 61/96 | Resistance regulation |
| SLUN_RS38460 WP_108155196.1 | Lun11 342 | StenM, N-acetyl-gamma-glutamyl-phosphate reductase (ArgC), stenothricin BGC from Streptomyces roseosporus (EFE73305.1) | 81/100 | L-ornithine synthesis |
| SLUN_RS38465 WP_108155197.1 | Lun12 417 | StenJ, N(2)-acetyl-L-ornithine:2-oxoglutarate aminotransferase (ArgD), stenothricin BGC from Streptomyces roseosporus NRRL 15998 (EFE73302.1) | 72/94 | L-ornithine synthesis |
| SLUN_RS38470 WP_108155198.1 | Lun13 300 | StenK, N-acetylglutamate kinase (ArgB), stenothricin BGC from Streptomyces roseosporus (EFE73303.1) | 81/97 | L-ornithine synthesis |
| SLUN_RS38475 WP_108155199.1 | Lun14 384 | StenL, bifunctional N(2)-acetyl-L-ornithine:L-glutamate N-acetyltransferase (ArgJ), stenothricin BGC from Streptomyces roseosporus (EFE73304.1) | 80/100 | L-ornithine synthesis |
| SLUN_RS38480 WP_108155200.1 | Lun15 70 | KtzJ, aurantimycin A BGC from Streptomyces aurantiacus JA 4570 (WP_016638464.1) | 71/100 | Building block modification |
| SLUN_RS38485 WP_108155201.1 | Lun16 444 | HmtM, lysine N(6)-hydroxylase/L-ornithine N(5)-oxygenase, himastatin BGC of Streptomyces himastatinicus (CBZ42147.1) | 63/97 | Piz synthesis |
| SLUN_RS38490 WP_108155202.1 | Lun17 230 | HmtC, piperazate synthase, himastatin BGC Streptomyces himastatinicus (CBZ42137.1) | 69/95 | Piz synthesis |
| SLUN_RS38495 WP_108155203.1 | Lun18 235 | LmbU family transcriptional regulator, himastatin BGC Streptomyces himastatinicus (CBZ42138.1) | 69/95 | Regulation |
| SLUN_RS38500 WP_108155204.1 | Lun19 100 | chorismate mutase, no hit in MiBIG, Streptomyces sp. SLBN-118 (WP_142214144.1) | 78/97 | Phe biosynthetic pathway |
| SLUN_RS38505 WP_108155205.1 | Lun20 1072 | OciC, non-ribosomal peptide synthetase (Module 3, domain C-A-T), cyanopeptolin BGC from Planktothrix agardhii NIVA-CYA 116 (ABI26079.1) | 43/98 | Incorporated aa: L-Ile |
| SLUN_RS38510 WP_108155206.1 | Lun21 400 | NAD(P)/FAD-dependent oxidoreductase, aurantimycin A BGC from Streptomyces aurantiacus JA 4570 (WP_016638468.1) | 53/99 | Hydroxylation |
| SLUN_RS38515 WP_108155207.1 | Lun22 5462 | HmtL, non-ribosomal peptide synthetase (Modules 4/5/6/7, domains C-A-T-E/C-A-T/C-A-T-E/C-A-T-TE), himastatin BGC of Streptomyces himastatinicus (CBZ42146.1) | 55–100 | Incorporated aa: D-Piz/L-Piz/D-Piz/L-Piz and peptide release and cyclization |
| SLUN_RS38520 WP_159100458.1 | Lun23 2167 | Non-ribosomal peptide synthetase (Module 1/2, domains A-T-E/C-A-T), aurantimycin A BGC from Streptomyces aurantiacus JA 4570 (WP_016638470.1) | 53/100 | Incorporated aa: D-Phe/inactive |
| SLUN_RS38525 WP_108155209.1 | Lun24 259 | KtzF thioesterase, alpha/beta fold hydrolase, himastatin BGC of Streptomyces himastatinicus (CBZ42144.1) | 58/97 | Peptide release |
| SLUN_RS38530 WP_108155210.1 | Lun25 258 | ABC transporter permease, aurantimycin A BGC from Streptomyces aurantiacus JA 4570 (WP_016638474.1) | 51/100 | Drug resistance |
| SLUN_RS38535 WP_108155211.1 | Lun26 323 | Daunorubicin resistance protein DrrA family ABC transporter ATP-binding protein, aurantimycin A BGC of Streptomyces aurantiacus JA 4570 (WP_016638473.1) | 66/94 | Drug resistance |
| SLUN_RS38540 WP_108155212.1 | Lun27 286 | DUF4097 family beta strand repeat-containing protein, himastatin BGC Streptomyces himastatinicus (CBZ42150.1) | 39/100 | Unknown |
| SLUN_RS38545 WP_108155213.1 | Lun28 261 | 3-oxoacyl-ACP reductase FabG, kiamycin biosynthetic gene cluster from Streptomyces sp. W007 (EHM27508.1) | 74/97 | Unknown |
| SLUN_RS38550 WP_108155214.1 | Lun29 285 | SARP family transcriptional regulator, mitomycin biosynthetic gene cluster from Streptomyces lavendulae (AAD28451.1) | 30/90 | Regulation |
| SLUN_RS38555 WP_108155215.1 | Lun30 293 | class I SAM-dependent methyltransferase, 5-isoprenylindole-3-carboxylate β-D-glycosyl ester BGC from Streptomyces sp. RM-5–8 (ANA09433.1) | 31/85 | Unknown |
| SLUN_RS38560 WP_108155216.1 | Lun31 108 | ArsR family transcriptional regulator, no hit in MiBIG database, Streptomyces mangrovisoli (WP_052743136.1) | 93/85 | Regulation |
| SLUN_RS38565 WP_108155217.1 | Lun32 457 | M14 family zinc carboxypeptidase, capreomycin IA BGC from Saccharothrix mutabilis subsp. Capreolus (ABR67765.1) | 45/98 | Unknown |
| SLUN_RS38570 WP_108155218.1 | Lun33 500 | Amino acid permease, amphotericin B biosynthetic gene cluster from Streptomyces nodosus (AAV48836.1) | 54/89 | Unkown |
| SLUN_RS38575 WP_257153969.1 | Lun34 450 | glutamine synthetase family protein, colabomycin E BGC from Streptomyces aureus (AIL50155.1) | 36/100 | Unknown |
| SLUN_RS38580 WP_108155220.1 | Lun35 190 | MarR family transcriptional regulator, no hit in MiBIG database, Streptomyces sp. ISL-44 (WP_215144797.1) | 83/93 | Regulation |
| SLUN_RS38585 WP_108155221.1 | Lun36 268 | acetoacetate decarboxylase family protein, enduracidin BGC from Streptomyces fungicidicus (ABD65946.1) | 35/99 | Unknown |
| SLUN_RS38590 WP_108155222.1 | Lun37 846 | SpoIIE family protein phosphatase, laidlomycin BGC from Streptomyces sp. CS684 (AFL48546.1) | 48/68 | Unknown |
| SLUN_RS38595 WP_108155223.1 | Lun38 256 | gamma-glutamyl-gamma-aminobutyrate hydrolase, nenestatin biosynthetic gene cluster from Micromonospora echinospora (ARD70860.1) | 35/95 | Unknown |
| SLUN_RS41955 WP_108155224.1 | Lun39 495 | Putative aldehyde dehydrogenase, pederin BGC from uncultured bacterium (AAS47555.1) | 49/100 | Unknown |
| Atom Number | Major Compound (1) | Minor Compound (2) | ||
|---|---|---|---|---|
| 1H NMR a | 13C NMR b | 1H NMR a | 13C NMR b | |
| 1 | 9.27 (b, 1H) | 9.64 (b, 1H) | ||
| 2 | 5.82–5.76 | 56.4 | 5.96 (d, J = 11.0 Hz, 1H) | 55.6 |
| 3 | 2.07–2.03 | 32.8 | 2.12–2.07 | 32.3 |
| 4 | 1.02–0.94 | 25.2 | 1.02–0.94 | 25.2 |
| 5 | 0.82–0.76 | 11.1 | 0.82–0.76 | 10.9 |
| 6 | 172.7 | 172.2 | ||
| 7 | 0.88–0.84 | 15.5 | 0.88–0.84 | 15.4 |
| 8 | 4.92–4.84 c | 4.92–4.84 c | ||
| 9 | 2.9–3.1 d | 46.9 d | e | e |
| 10 | 1.2–2.3 f | 18.4–26.2 f | e | e |
| 11 | 1.2–2.3 f | 18.4–26.2 f | e | e |
| 12 | 5.75 (d, J = 5.6 Hz, 1H) g | 48.9 g | 5.82–5.76 g | 47.6 g |
| 13 | 173.5 h | 171.3 h | ||
| 14 | 5.05 (d, J = 13.8 Hz, 1H) c | |||
| 15 | 2.9–3.1 d | 47.1 d | 6.91 (d, J = 3.7 Hz, 1H) i | 142.9 i |
| 16 | 1.2–2.3 f | 18.4–26.2 f | e | e |
| 17 | 1.2–2.3 f | 18.4–26.2 f | e | e |
| 18 | 5.39 g | 48.1 g | 5.36–5.28 g | 47.5 g |
| 19 | 174.1 h | 172.9 h | ||
| 20 | 5.18 (d, J = 13.0 Hz, 1H) c | 5.01 (d, J = 14.4 Hz, 1H) c | ||
| 21 | 2.9–3.1 d | 47.2 d | e | e |
| 22 | 1.2–2.3 f | 18.4–26.2 f | e | e |
| 23 | 1.2–2.3 f | 18.4–26.2 f | e | e |
| 24 | 5.45 g | 47.6 g | 5.67 g | 46.7 g |
| 25 | 173.2 h | 171.6 h | ||
| 26 | 5.23 (dd, J = 12.8, 3.4 Hz, 1H) c | 5.36–5.28 c | ||
| 27 | 2.9–3.1 d | 47.3 d | e | e |
| 28 | 1.2–2.3 f | 18.4–26.2 f | e | e |
| 29 | 1.2–2.3 f | 18.4–26.2 f | e | e |
| 30 | 4.92–4.84 g | 49.6 g | 5.37 g | 47.6 g |
| 31 | 170.9(8)h | 171.0(2) h | ||
| 32 | 8.27 (d, J = 8.6 Hz, 1H) | 8.22 (d, J = 9.0 Hz, 1H) | ||
| 33 | 5.36–5.28 | 49.0 | 5.68 (d, J = 5.4 Hz, 1H) | 49.1 |
| 34 | 2.93–2.86 and 2.77–2.67 | 37.7 | 2.93–2.86 and 2.77–2.67 | 37.7 |
| 35 | 137.7(1) | 137.7(4) | ||
| 36 | 7.20–7.14 j | 129.7(0) | 7.20–7.14 j | 129.7(3) |
| 37 | 7.24 j | 128.4 | 7.24 j | 128.3 |
| 38 | 7.20–7.14 j | 126.6(7) | 7.20–7.14 j | 126.6(5) |
| 39 | 7.24 j | 128.4 | 7.24 j | 128.3 |
| 40 | 7.20–7.14 j | 129.7(0) | 7.20–7.14 j | 129.7(3) |
| 41 | 170.3 | 170.4 | ||
| Nb | RT (min) | Name | m/z Observed [M+H]+/[M+Na]+ | Formula [M+H]+/[M+Na]+ | Δm (ppm) |
|---|---|---|---|---|---|
| 1 | 18.35 | Lunaemycin A | 725.4092/747.3903 | C35H53N10O7+/C35H52N10O7Na+ | 0.1/1 |
| 2a | 17.42 | Lunaemycin B1 | 723.3935/745.3749 | C35H51N10O7+/C35H50N10O7Na+ | 0.2/0.9 |
| 2b | 17.74 | Lunaemycin B2 | 723.3935/745.3749 | C35H51N10O7+/C35H50N10O7Na+ | 0.2/0.9 |
| 3a | 16.94 | Lunaemycin C1 | 721.3779/743.3594 | C35H49N10O7+/C35H48N10O7Na+ | 0.1/0.4 |
| 3b | 17.27 | Lunaemycin C2 | 721.3777/743.3594 | C35H49N10O7+/C35H48N10O7Na+ | 0.4/0.8 |
| 4 | 16.82 | Lunaemycin D | 739.3886/761.3698 | C35H51N10O8+/C35H50N10O8Na+ | 0.1/1 |
| 5a | 15.91 | Lunaemycin E1 | 737.3731/759.3546 | C35H49N10O8+/C35H48N10O8Na+ | 0.2/0.4 |
| 5b | 16.44 | Lunaemycin E2 | 737.3731/759.3546 | C35H49N10O8+/C35H48N10O8Na+ | 0.6/1.3 |
| 6 | 15.60 | Lunaemycin F | 741.4042/763.3856 | C35H53N10O8+/C35H52N10O8Na+ | 0.1/0.7 |
| 7 | 13.76 | Lunaemycin G | 755.3832/777.3647 | C35H51N10O9+/C35H50N10O9Na+ | 0.3/0.9 |
| 8 | 12.81 | Lunaemycin H | 757.399/779.3806 | C35H53N10O9+/C35H52N10O7Na+ | 0.1/0.3 |
| 9 | 17.59 | Lunaemycin I | 711.3935/733.3751 | C34H51N10O7+/C34H48N10O7Na+ | 0.1/0.8 |
| 10a | 16.70 | Lunaemycin J1 | 709.3784/731.3603 | C34H49N10O7+/C34H48N10O7Na+ | 0.3/0.3 |
| 10b | 17. 10 | Lunaemycin J2 | 709.3777/731.3594 | C34H49N10O7+/C34H48N10O7Na+ | 0.3/0.5 |
| 10c | 17.40 | Lunaemycin J3 | 709.4141/731.3951 | C35H53N10O7+/C35H52N10O6Na+ | 0.3/0.1 |
| Additional lunaemycin derivatives with non-elucidated structure | |||||
| 11 | 11.73 | - | 753.3666/775.3492 | C35H49N10O9+/C35H48N10O9Na+ | 1.7/0.8 |
| 12 | 11.79 | - | 771.3785/793.36 | C35H51N10O10+/C35H50N10O10Na+ | 0.2/0.5 |
| 13 | 15.91 | - | 754.3993 | C35H52N11O8+/ND | 0.3 |
| 14 | 16.84 | - | 613.346 | C30H45N8O6+/ND | 0.4 |
| 15 | 18.33 | - | 742.436 | C35H56N10O7+/ND | 1.2 |
| The numbers attributed to the different lunaemycins refer to the peaks displayed in EIC of Figure 7. | |||||
| Organism | Resistance | MIC (µg/mL) | MBC (µg/mL) | Comparator (MIC, µg/mL) a |
|---|---|---|---|---|
| Gram-positive bacteria | ||||
| Bacillus subtilis ATCC 6633 | - | 0.12 | 4 | Van, 0.5 |
| Enterococcus faecalis ATCC 29212 | - | 0.12 | 8 | Van, 1 |
| Streptococcus pyogenes ATCC 12344 | - | 0.12 | 8 | Van, 1 |
| Staphylococcus aureus ATCC 25923 | - | 0.12 | 4 | Van, 0.5 |
| Enterococcus faecalis SI-759 | VancomycinR, teicoplaninR | 0.25 | 8 | Tei, 4 |
| Enterococcus faecium SI-1831 | VancomycinR, teicoplaninR | 0.25 | 0.25 | Tei, 32 |
| Staphylococcus aureus ATCC 43300 | MethicillinR, MRSA | 0.12 | 2 | Oxa, 32 |
| Staphylococcus epidermidis SI-1266 | LinezolidR | 0.12 | 0.25 | Lin, 64 |
| Staphylococcus haemolyticus SI-6/2011 | GentamicinR | 0.12 | 2 | Gen, >32 |
| Staphylococcus warneri SI-5/2011 | OxacillinR | 0.12 | 8 | Oxa, 32 |
| Gram-negative bacteria | ||||
| Acinetobacter baumannii ATCC 17978 | - | >64 | - | Col, 1 |
| Escherichia coli CCUGT | - | >64 | - | Col, 0.5 |
| Klebsiella pneumoniae ATCC 13833 | - | >64 | - | Col, 0.5 |
| Pseudomonas aeruginosa ATCC 27853 | - | >64 | - | Col, 1 |
| Acinetobacter baumannii N50 | MDR, carbapenemR | >64 | - | Mer, 32 |
| Escherichia coli SI-63 | MDR, colistinR, mcr+ | >64 | - | Col, 8 |
| Escherichia coli MO287 | MDR, NDM-1+ | >64 | - | Mer, 64 |
| Escherichia coli SI-1896 | MDR, OXA-48-like+ | >64 | - | Mer, >16 |
| Klebsiella pneumoniae B2 | pandrug-resistant | >64 | - | Col, 128 |
| Klebsiella pneumoniae BO4 | pandrug-resistant | >64 | - | Col, 64 |
| Klebsiella pneumoniae SI-518 | MDR, NDM-1+ | >64 | - | Mer, 128 |
| Klebsiella pneumoniae SI-703A | MDR, OXA-48-like+ | >64 | - | Mer, >16 |
| Pseudomonas aeruginosa SI-270 | MDR, colistinR | >64 | - | Col, 16 |
| Bacteria | Ref Number | Comment/Utilization | Origin |
|---|---|---|---|
| Streptomyces Strains | |||
| S. lunaelactis MM109T | DSM 42149 | BGC 28a in pSLUN1a, type strain. Lunaemycin producer | ULiège/Hedera 22 |
| S. lunaelactis MM37 | - | BGC 28a in pSLUN1a. Lunaemycin producer | ULiège/Hedera 22 |
| S. lunaelactis MM22 | - | BGC 28b in pSLUN1b | ULiège/Hedera 22 |
| S. lunaelactis MM25 | - | BGC 28b in pSLUN1b | ULiège/Hedera 22 |
| S. lunaelactis MM28 | - | BGC 28b in pSLUN1b | ULiège/Hedera 22 |
| S. lunaelactis MM31 | - | BGC 28b in pSLUN1b. | ULiège/Hedera 22 |
| S. lunaelactis MM40 | - | BGC 28b in pSLUN1b | ULiège/Hedera 22 |
| S. lunaelactis MM78 | - | BGC 28b in pSLUN1b | ULiège/Hedera 22 |
| S. lunaelactis MM83 | - | BGC 28b in pSLUN1b | ULiège/Hedera 22 |
| S. lunaelactis MM113 | - | BGC 28b in pSLUN1b | ULiège/Hedera 22 |
| S. lunaelactis MM115 | - | BGC 28b in pSLUN1b | ULiège/Hedera 22 |
| Strains used for antibacterial assays | |||
| Acinetobacter baumannii | ATCC 17978 | Reference strain | ULiège |
| Acinetobacter baumannii | N50 | Clinical isolate (MDR, carbapnemR) | Università di Siena |
| Bacillus subtilis | ATCC 6633 | Reference strain | Università di Siena |
| Bacillus subtilis | ATCC 19659 | Reference strain | ULiège |
| Citrobacter freundii | ATCC 43864 | Reference strain | ULiège |
| Escherichia coli | CCUGT | Reference strain | Università di Siena |
| Escherichia coli | ATCC 25922 | Reference strain | ULiège |
| Escherichia coli | SI-63 | Clinical isolate (MDR, colistinR, mcr+) | Università di Siena |
| Escherichia coli | MO287 | Clinical isolate (MDR, NDM-1+) | Università di Siena |
| Escherichia coli | SI-1896 | Clinical isolate (MDR, OXA-48-like+) | Università di Siena |
| Enterococcus faecalis | ATCC 29212 | Reference strain | Università di Siena |
| Enterococcus faecalis | SI-759 | Clinical isolate (vancomycinR, teicoplaninR) | Università di Siena |
| Enterococcus faecium | SI-1831 | Clinical isolate (vancomycinR, teicoplaninR) | Università di Siena |
| Enterococcus hirae | ATCC 9790 | Reference strain | ULiège |
| Klebsiella pneumoniae | ATCC 13833 | Reference strain | ULiège/Università di Siena |
| Klebsiella pneumoniae | B2 | Clinical isolate (pandrug-resistant) | Università di Siena |
| Klebsiella pneumoniae | BO4 | Clinical isolate (pandrug-resistant) | Università di Siena |
| Klebsiella pneumoniae | SI-518 | Clinical isolate (MDR, NDM-1+) | Università di Siena |
| Klebsiella pneumoniae | SI-703A | Clinical isolate (MDR, OXA-48-like+) | Università di Siena |
| Kocuria rhizophila (Micrococcus luteus) | ATCC 9341 | Reference strain | ULiège |
| Proteus mirabilis | ATCC 7002 | Reference strain | ULiège |
| Pseudomonas aeruginosa | ATCC 27853 | Reference strain | ULiège/Università di Siena |
| Pseudomonas aeruginosa | SI-270 | Clinical isolate (MDR, colistinR) | Università di Siena |
| Serratia marcescens | ATCC 10759 | Reference strain | ULiège |
| Streptococcus pyogenes | ATCC 12344 | Reference strain | ULiège |
| Staphylococcus aureus | ATCC 25923 | Reference strain | ULiège/Università di Siena |
| Staphylococcus aureus | ATCC 43300 | Clinical isolate (methicillinR, MRSA) | Università di Siena |
| Staphylococcus epidermidis | SI-1266 | Clinical isolate (linezolidR) | Università di Siena |
| Staphylococcus haemolyticus | SI-6/2011 | Clinical isolate (gentamicinR) | Università di Siena |
| Staphylococcus warneri | SI-5/2011 | Clinical isolate (oxacillinR) | Università di Siena |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinet, L.; Naômé, A.; Rezende, L.C.D.; Tellatin, D.; Pignon, B.; Docquier, J.-D.; Sannio, F.; Baiwir, D.; Mazzucchelli, G.; Frédérich, M.; et al. Lunaemycins, New Cyclic Hexapeptide Antibiotics from the Cave Moonmilk-Dweller Streptomyces lunaelactis MM109T. Int. J. Mol. Sci. 2023, 24, 1114. https://doi.org/10.3390/ijms24021114
Martinet L, Naômé A, Rezende LCD, Tellatin D, Pignon B, Docquier J-D, Sannio F, Baiwir D, Mazzucchelli G, Frédérich M, et al. Lunaemycins, New Cyclic Hexapeptide Antibiotics from the Cave Moonmilk-Dweller Streptomyces lunaelactis MM109T. International Journal of Molecular Sciences. 2023; 24(2):1114. https://doi.org/10.3390/ijms24021114
Chicago/Turabian StyleMartinet, Loïc, Aymeric Naômé, Lucas C. D. Rezende, Déborah Tellatin, Bernard Pignon, Jean-Denis Docquier, Filomena Sannio, Dominique Baiwir, Gabriel Mazzucchelli, Michel Frédérich, and et al. 2023. "Lunaemycins, New Cyclic Hexapeptide Antibiotics from the Cave Moonmilk-Dweller Streptomyces lunaelactis MM109T" International Journal of Molecular Sciences 24, no. 2: 1114. https://doi.org/10.3390/ijms24021114
APA StyleMartinet, L., Naômé, A., Rezende, L. C. D., Tellatin, D., Pignon, B., Docquier, J. -D., Sannio, F., Baiwir, D., Mazzucchelli, G., Frédérich, M., & Rigali, S. (2023). Lunaemycins, New Cyclic Hexapeptide Antibiotics from the Cave Moonmilk-Dweller Streptomyces lunaelactis MM109T. International Journal of Molecular Sciences, 24(2), 1114. https://doi.org/10.3390/ijms24021114