High Duty Cycle Echolocation May Constrain the Evolution of Diversity within Horseshoe Bats (Family: Rhinolophidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Animals
2.2. Sampling
2.3. Phylogenetic Analyses
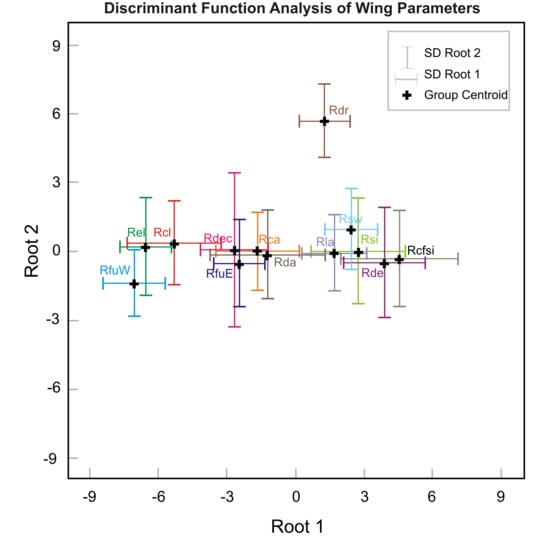
2.4. Wing Morphology
2.5. Echolocation
2.6. Functional Association between Echolocation and Wing Morphology
2.7. Sexual Dimorphism
2.8. Statistical Analyses
3. Results
3.1. Phylogeny
3.2. Functional Association between Body Size and Wing Morphology
3.3. Functional Association between Body Size and Echolocation among Species
3.4. Phylogenetically Informed Analyses of Associations between Body Size, Wing Morphology and Echolocation among Species
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jezkova, T.; Wiens, J.J. What explains patterns of diversification and richness among animal phyla? Am. Nat. 2017, 189, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.M.; Wheat, C.W.; Heckel, D.G.; Vogel, H. Evolutionary origins of a novel host plant detoxification gene in butterflies. Mol. Biol. Evol. 2008, 25, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.J.; Matos-Maraví, P.F.; Beheregaray, L.B. Delving into Delias Hübner (Lepidoptera: Pieridae): Fine-scale biogeography, phylogenetics and systematics of the world's largest butterfly genus. J. Biogeogr. 2013, 40, 881–893. [Google Scholar] [CrossRef]
- Simmons, N.B. Order Chiroptera. In Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed.; Wilson, D.E., Reeder, D.M., Eds.; Bucknell University: Lewisburg, PA, USA, 2005; pp. 312–529. [Google Scholar]
- Futuyma, D.J. Evolutionary Biology; Sinauer Associates: Sunderland, MA, USA, 1998. [Google Scholar]
- Roff, D.A.; Fairbairn, D.J. The evolution of trade-offs: Where are we? J. Evol. Biol. 2007, 20, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, D.G.; Evans, H.E. Why are there no viviparous birds? Am. Nat. 1986, 128, 165–190. [Google Scholar] [CrossRef]
- Jacobs, D.S.; Babiker, H.; Bastian, A.; Kearney, T.; van Eeden, R.; Bishop, J.M. Phenotypic convergence in genetically distinct lineages of a Rhinolophus species complex (mammalia, chiroptera). PLoS ONE 2013, 8, e82614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobs, D.S.; Bastian, A.; Bam, L. The influence of feeding on the evolution of sensory signals: A comparative test of an evolutionary trade-off between masticatory and sensory functions of skulls in southern African horseshoe bats (Rhinolophidae). J. Evol. Biol. 2014, 27, 2829–2840. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, H.; Rautenbach, I. Morphology, echolocation and resource partitioning in insectivorous bats. J. Anim. Ecol. 1987, 56, 763–778. [Google Scholar] [CrossRef]
- Norberg, U.M.; Rayner, J.M.V. Ecological morphology and flight in bats (Mammalia; Chiroptera): Wing adaptations, flight performance, foraging strategy and echolocation. Phil. Trans. R. Soc. B Biol. Sci. 1987, 316, 335–427. [Google Scholar] [CrossRef]
- Jacobs, D.S.; Barclay, R.M.; Walker, M.H. The allometry of echolocation call frequencies of insectivorous bats: Why do some species deviate from the pattern? Oecologia 2007, 152, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.A.; Moss, C.F.; Vater, M. Echolocation in Bats and Dolphins; University of Chicago Press: Chicago, IL, USA, 2004. [Google Scholar]
- Jones, G.; Siemers, B.M. The communicative potential of bat echolocation pulses. J. Comp. Physiol. A 2011, 197, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Fenton, M.B. Describing the echolocation calls and behaviour of bats. Acta Chiropterol. 1999, 1, 127–136. [Google Scholar]
- Schnitzler, H.-U.; Flieger, E. Detection of oscillating target movements by echolocation in the Greater horseshoe bat. J. Comp. Physiol. A 1983, 153, 385–391. [Google Scholar] [CrossRef]
- Neuweiler, G. Foraging, echolocation and audition in bats. Naturwissenschaften 1984, 71, 446–455. [Google Scholar] [CrossRef]
- Schnitzler, H.-U.; Denzinger, A. Auditory fovea and Doppler shift compensation: Adaptations for flutter detection in echolocating bats using CF-FM signals. J. Comp. Physiol. A 2011, 197, 541–559. [Google Scholar] [CrossRef] [PubMed]
- Schnitzler, H.-U.; Kalko, E.K.V. Echolocation by insect-eating bats. BioScience 2001, 51, 557–569. [Google Scholar] [CrossRef]
- Lawrence, B.D.; Simmons, J.A. Measurements of atmospheric attenuation at ultrasound frequencies and the significance for echolocation by bats. J. Acoust. Soc. Am. 1982, 71, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Denzinger, A.; Schnitzler, H.-U. Bat guilds, a concept to classify the highly diverse foraging and echolocation behaviors of microchiropteran bats. Front. Physiol. 2013, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Csorba, G.; Ujhelyi, P.; Thomas, N. Horseshoe Bats of the World (Chiroptera: Rhinolophidae); Alana Books: Shropshire, UK, 2003. [Google Scholar]
- Dool, S.E.; Puechmaille, S.J.; Foley, N.M.; Allegrini, B.; Bastian, A.; Mutumi, G.L.; Maluleke, T.G.; Odendaal, L.J.; Teeling, E.C.; Jacobs, D.S. Nuclear introns outperform mitochondrial DNA in inter-specific phylogenetic reconstruction: Lessons from horseshoe bats (Rhinolophidae: Chiroptera). Mol. Phylogenet. Evol. 2016, 97, 196–212. [Google Scholar] [CrossRef] [PubMed]
- Ammerman, L.K.; Lee, D.N.; Tipps, T.M. First molecular phylogenetic insights into the evolution of free-tailed bats in the subfamily Molossinae (Molossidae, Chiroptera). J. Mammal. 2012, 93, 12–28. [Google Scholar] [CrossRef] [Green Version]
- Fenton, M.B.; Bogdanowicz, W. Relationships between external morphology and foraging behaviour: Bats in the genus Myotis. Can. J. Zool. 2002, 80, 1004–1013. [Google Scholar] [CrossRef]
- Ruedi, M.; Mayer, F. Molecular systematics of bats of the genus Myotis (Vespertilionidae) suggests deterministic ecomorphological convergences. Mol. Phylogenet. Evol. 2001, 21, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, D.S.; Mutumi, G.L.; Maluleke, T.; Webala, P.W. Convergence as an evolutionary trade-off in the evolution of acoustic signals: Echolocation in horseshoe bats as a case study. In Evolutionary Biology; Springer: New York, NY, USA, 2016; pp. 89–103. [Google Scholar]
- Fairbairn, D.J. Allometry for sexual size dimorphism: Pattern and process in the coevolution of body size in males and females. Annu. Rev. Ecol. Syst. 1997, 28, 659–687. [Google Scholar] [CrossRef]
- Dale, J.; Dunn, P.O.; Figuerola, J.; Lislevand, T.; Székely, T.; Whittingham, L.A. Sexual selection explains Rensch’s rule of allometry for sexual size dimorphism. Proc. R. Soc. B Biol. Sci. 2007, 274, 2971–2979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoffberg, S.; Jacobs, D.S.; Matthee, C.A. The divergence of echolocation frequency in horseshoe bats: Moth hearing, body size or habitat? J. Mamm. Evol. 2011, 18, 117–129. [Google Scholar] [CrossRef]
- Foley, N.M.; Thong, V.D.; Soisook, P.; Goodman, S.M.; Armstrong, K.N.; Jacobs, D.S.; Puechmaille, S.J.; Teeling, E.C. How and why overcome the impediments to resolution: Lessons from rhinolophid and hipposiderid bats. Mol. Biol. Evol. 2014, 32, 313–333. [Google Scholar] [CrossRef] [PubMed]
- Cotterill, F.P.; Happold, M. Rhinolophus darlingi darling’s horseshoe bat. In Mammals of Africa Volume III: Rodents, Hares and Rabbits; Bloomsbury: New York, NY, USA, 2013. [Google Scholar]
- Monadjem, A.; Taylor, P.J.; Cotterill, W.; Schoeman, M.C. Bats of Southern and Central Africa: A Biogeographic and Taxonomic Synthesis, 1st ed.; Wits University Press: Johannesburg, South Africa, 2010. [Google Scholar]
- Racey, P. Ageing and assessment of reproductive status of pipistrelle bats, Pipistrellus pipistrellus. J. Zool. 1974, 173, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Anthony, E. Age determination in bats. Ecological and Behavioral Methods for the Study of Bats; Smithsonian Institution Press: Washington, DC, USA, 1988; pp. 47–58. [Google Scholar]
- Rughetti, M.; Toffoli, R. Sex-specific seasonal change in body mass in two species of vespertilionid bats. Acta Chiropterol. 2014, 16, 149–155. [Google Scholar] [CrossRef]
- Puechmaille, S.J.; Gouilh, M.A.; Piyapan, P.; Yokubol, M.; Mie, K.M.; Bates, P.J.; Satasook, C.; Nwe, T.; Bu, S.S.; Mackie, I.J.; et al. The evolution of sensory divergence in the context of limited gene flow in the Bumblebee bat. Nat. Commun. 2011, 2, 573. [Google Scholar] [CrossRef] [PubMed]
- Igea, J.; Juste, J.; Castresana, J. Novel intron markers to study the phylogeny of closely related mammalian species. BMC Evol. Biol. 2010, 10, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salicini, I.; Ibáñez, C.; Juste, J. Multilocus phylogeny and species delimitation within the Natterer’s bat species complex in the western palearctic. Mol. Phylogenet. Evol. 2011, 61, 888–898. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.-I.; Miyata, T. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. Jmodeltest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the Cipres Science Gateway for Inference of Large Phylogenetic Trees. In Gateway Computing Environments Workshop (GCE); Institute of Electrical and Electronics Engineers: Piscataway, NJ, USA, 2010; pp. 1–8. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. Mrbayes: Bayesian inference of phylogeny. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.; Suchard, M. Tracer v1. 6—Mcmc Trace Analysis Package; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2013. [Google Scholar]
- Drummond, A.J.; Rambaut, A. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A. Figtree v1. 4. In Molecular Evolution, Phylogenetics and Epidemiology; University of Edinburgh, Institute of Evolutionary Biology: Edinburgh, UK, 2012. [Google Scholar]
- Paradis, E.; Bolker, B.; Claude, J.; Cuong, H.S.; Desper, R.; Legendre, J.L.; Noel, Y.; Nylander, J.; Opgen-Rhein, R.; Popescu, A.-A. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2012, 20, 289–290. [Google Scholar] [CrossRef]
- Saunders, M.B.; Barclay, R.M.R. Ecomorphology of insectivorous bats: A test of predictions using two morphologically similar species. Ethology 1992, 73, 1335–1345. [Google Scholar] [CrossRef]
- Siemers, B.M.; Beedholm, K.; Dietz, C.; Dietz, I.; Ivanova, T. Is species identity, sex, age or individual quality conveyed by echolocation call frequency in european horseshoe bats? Acta Chiropterol. 2005, 7, 259–274. [Google Scholar] [CrossRef]
- Schnitzler, H.-U. Echoes of fluttering insects: Information for echolocating bats. In Recent Advances in the Study of Bats; Fenton, B., Racey, P.A., Rayner, J.M.V., Eds.; Cambridge University Press: Cambridge, UK, 1987. [Google Scholar]
- Bastian, A.; Jacobs, D.S. Listening carefully: Increased perceptual acuity for species discrimination in multispecies signalling assemblages. Anim. Behav. 2015, 101, 141–154. [Google Scholar] [CrossRef]
- Jacobs, D.S. Evolution’s Chimera: Bats and the Marvel of Evolutionary Adaptation, 1st ed.; University of Cape Town Press: Cape Town, South Africa, 2016; p. 196. [Google Scholar]
- Dietz, C.; Dietz, I.; Siemers, B.M. Wing measurement variations in the five european horseshoe bat species (Chiroptera : Rhinolophidae). J. Mammal. 2006, 87, 1241–1251. [Google Scholar] [CrossRef]
- Smith, R.J. Statistics of sexual size dimorphism. J. Hum. Evol. 1999, 36, 423–458. [Google Scholar] [CrossRef] [PubMed]
- Webb, T.J.; Freckleton, R.P. Only half right: Species with female-biased sexual size dimorphism consistently break Rensch’s rule. PLoS ONE 2007, 2, e897. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, H.F. The application of electronic computers to factor analysis. Educ. Psychol. Meas. 1960, 20, 141–151. [Google Scholar] [CrossRef]
- Garamszegi, L.Z. Modern Phylogenetic Comparative Methods and Their Application in Evolutionary Biology; Springer: New York, NY, USA, 2014. [Google Scholar]
- Grafen, A. The phylogenetic regression. Philos. Trans. R. Soc. B Biol. Sci. 1989, 326, 119–157. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, version 3.5.0; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- R Studio Team. R Studio: Integrated Development Environment for R, version 0.99.463; R Studio Team: Boston, MA, USA, 2015. [Google Scholar]
- Orme, D.; Freckleton, R.; Thomas, G.; Petzoldt, T.; Fritz, S.; Isaac, N.; Pearse, W. Caper: Comparative Analyses of Phylogenetics and Evolution in R. Version 1.0.1. 2013. Available online: http://cran.r-project.org/web/packages/caper/vignettes/caper.pdf (accessed on 17 April 2018).
- Pagel, M. Inferring evolutionary processes from phylogenies. Zool. Scr. 1997, 26, 331–348. [Google Scholar] [CrossRef]
- Pagel, M. Inferring the historical patterns of biological evolution. Nature 1999, 401, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Harmon, L.J.; Weir, J.T.; Brock, C.D.; Glor, R.E.; Challenger, W. Geiger: Investigating evolutionary radiations. Bioinformatics 2007, 24, 129–131. [Google Scholar] [CrossRef] [PubMed]
- Freckleton, R.P.; Harvey, P.H.; Pagel, M. Phylogenetic analysis and comparative data: A test and review of evidence. Am. Nat. 2002, 160, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Stöver, B.C.; Müller, K.F. Treegraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Stevens, R.D.; Platt, R.N. Patterns of secondary sexual size dimorphism in new world myotis and a test of Rensch’s rule. J. Mammal. 2015, 96, 1128–1134. [Google Scholar] [CrossRef]
- Wu, H.; Jiang, T.; Huang, X.; Feng, J. Patterns of sexual size dimorphism in horseshoe bats: Testing Rensch’s rule and potential causes. Sci. Rep. 2018, 8, 2616. [Google Scholar] [CrossRef] [PubMed]
- Ulian, C.M.V.; Rossi, M.N. Intraspecific variation in body size and sexual size dimorphism, and a test of Rensch’s rule in bats. Acta Zool. 2017, 98, 377–386. [Google Scholar] [CrossRef]
- Puechmaille, S.J.; Borissov, I.M.; Zsebok, S.; Allegrini, B.; Hizem, M.; Kuenzel, S.; Schuchmann, M.; Teeling, E.C.; Siemers, B.M. Female mate choice can drive the evolution of high frequency echolocation in bats: A case study with Rhinolophus mehelyi. PLoS ONE 2014, 9, e103452. [Google Scholar] [CrossRef] [PubMed]
- Pélissié, B.; Jarne, P.; David, P. Sexual selection without sexual dimorphism: Bateman gradients in a simultaneous hermaphrodite. Evol. Int. J. Org. Evol. 2012, 66, 66–81. [Google Scholar] [CrossRef] [PubMed]
- Blanckenhorn, W.U. The evolution of body size: What keeps organisms small? Q. Rev. Biol. 2000, 75, 385–407. [Google Scholar] [CrossRef] [PubMed]
- Stearns, S.C. The Evolution of Life Histories; Oxford University Press: New York, NY, USA, 1992. [Google Scholar]
- James, F.C. Geographic size variation in birds and its relationship to climate. Ecology 1970, 51, 365–390. [Google Scholar] [CrossRef]
- Taylor, P.J.; Stoffberg, S.; Monadjem, A.; Schoeman, M.C.; Bayliss, J.; Cotterill, F.P.D. Four new bat species Rhinolophus hildebrandtii complex) reflect plio-pleistocene divergence of dwarfs and giants across an afromontane archipelago. PLoS ONE 2012, 7, e41744. [Google Scholar] [CrossRef] [PubMed]
- Heller, K.-G.; von Helversen, O. Resource partitioning of sonar frequency bands in rhinolophoid bats. Oecologia 1989, 80, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Novick, A. Acoustic Orientation. In Biology of Bats; Wimsatt, W.A., Ed.; Academic Press: New York, NY, USA; London, UK, 1977; Volume 3, pp. 73–287. [Google Scholar]
- Barclay, R.M.R.; Brigham, R.M. Prey detection, dietary niche breadth, and body size in bats: Why are aerial insectivorous bats so small? Am. Nat. 1991, 137, 693–703. [Google Scholar]
- Jones, G. Does Echolocation Constrain the Evolution of Body Size in Bats? Symposia of the Zoological Society of London: London, UK, 1996; pp. 111–128. [Google Scholar]
- Luo, J.; Koselj, K.; Zsebők, S.; Siemers, B.M.; Goerlitz, H.R. Global warming alters sound transmission: Differential impact on the prey detection ability of echolocating bats. J. R. Soc. Interface 2014, 11, 20130961. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, L.; Brinkløv, S.; Surlykke, A. Intensity and directionality of bat echolocation signals. Front. Physiol. 2013, 4, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, G. Scaling of wingbeat and echolocation pulse emission rates in bats: Why are aerial insectivorous bats so small? Funct. Ecol. 1994, 8, 450–457. [Google Scholar] [CrossRef]






| Species Comparison | lnWL × lnMass | logRF × logMass | logRF × logDur | ResidRF × ResidWL |
| Lambda (ML) | 0.00 | 0.00 | 0.56 | 0.00 |
| Intercept b (SE) | 0.98 (0.23) | 2.41 (0.14) | 2.10 (0.41) | 6.86 × 10−10 (2.39 × 10−2) |
| t-test | 4.30 | 17.52 | 5.18 | 0.00 |
| p-value | 0.001 * | 2.2 × 10−9 * | 0.000 * | 1.00 |
| Slope β (SE) | 0.41 (0.11) | −0.51 (0.13) | −0.13 (0.26) | −9.41 × 10−2 (1.79 × 10−1) |
| t-test | 4.18 | −3.76 | −0.51 | −0.53 |
| p-value | 0.002 * | 0.003 * | 0.62 | 0.61 |
| Adjusted R2 | 0.58 | 0.52 | −0.07 | 0.064 |
| F-ratio | 17.47 | 14.16 | 0.26 | 0.28 |
| p-value (df 1, 11) | 0.002 * | 0.003 * | 0.62 | 0.61 |
| Sexual Dimorphism | lnMassf × lnMassmf | lnFAf × lnFAmf | lnWSf × lnWSmf | |
| Lambda (ML) | 0.66 | 0.57 | 0.58 | |
| Intercept b (SE) | 2.29 (0.25) | 1.47 (0.07) | 3.45 (0.06) | |
| t-test | 9.28 | 21.14 | 61.06 | |
| p-value | 1.561 × 10−6 * | 2.951 × 10−10 * | 2.665 × 10−15 * | |
| Slope β (SE) | −1.12 (2.05) | −5.17 (2.25) | −0.72 (0.80) | |
| t-test | −0.60 | −2.30 | −0.90 | |
| p-value | 0.56 | 0.04 | 0.39 | |
| Adjusted R2 | −0.06 | 0.26 | −0.02 | |
| F-ratio | 0.36 | 5.28 | 0.82 | |
| p-value (df 1, 11) | 0.56 | 0.042 * | 0.39 |
| Variable | PCA Factor Loadings | ||||
| PC 1 | PC 2 | PC 3 | |||
| Mass | −0.97 | ||||
| FA | −0.97 | ||||
| Wing area | −0.91 | ||||
| Wingspan (cm) | −0.90 | ||||
| Wing loading (Nm-2) | −0.79 | ||||
| Wingtip Shape Index | −0.89 | ||||
| Wingtip area ratio | −0.85 | ||||
| Aspect ratio | −0.83 | ||||
| Eigenvalues | 4.22 | 1.86 | 1.14 | ||
| Cumulative % | 46.84 | 67.48 | 80.17 | ||
| Variable | DFA Results | ||||
| Root 1 | Root 2 | Wilks’ λ | F to Remove | p | |
| PC 1 | 0.96 | 0.23 | 0.08 | 839.91 | <0.0001 |
| PC 3 | −0.03 | 0.63 | 0.74 | 21.40 | <0.0001 |
| PC 2 | 0.01 | −0.60 | 0.76 | 24.07 | <0.0001 |
| Eigenvalue | 13.10 | 0.75 | |||
| Cumulative % | 94.1 | 99.5 | |||
| Wilks’ λ | 0.04 | 0.54 | |||
| χ2 | 2723.80 | 519.37 | |||
| df | 36 | 22 | |||
| p | <0.0001 | <0.0001 | |||
| Variable | PCA Factor Loadings | ||||
| PC 1 | PC 2 | PC 3 | PC 4 | ||
| Inter-onset interval | 0.85 | ||||
| Inter-pulse interval | 0.78 | ||||
| Duration of pulse | 0.75 | ||||
| Sweep rate of FMt | −0.74 | ||||
| Minimum frequency of FMt | 0.91 | ||||
| Resting frequency | 0.90 | ||||
| Minimum frequency of FMi | 0.86 | ||||
| Duration of FMt | −0.73 | ||||
| Sweep rate of FMi | 0.68 | ||||
| Duty cycle | −0.73 | ||||
| Eigenvalues | 4.08 | 3.12 | 1.97 | 1.49 | |
| Cumulative % | 34.01 | 59.98 | 76.42 | 88.82 | |
| Variable | DFA Results | ||||
| Root 1 | Root 2 | Wilks’ λ | F to Remove | p | |
| PC 2 | −0.33 | 0.59 | 0.14 | 282.60 | <0.0001 |
| PC 1 | 0.07 | 0.48 | 0.48 | 48.50 | <0.0001 |
| PC 3 | 0.05 | 0.58 | 0.49 | 46.53 | <0.0001 |
| PC 4 | 0.01 | −0.37 | 0.72 | 16.83 | <0.0001 |
| Eigenvalue | 54.87 | 1.38 | |||
| Cumulative % | 95.82 | 98.23 | |||
| Wilks’ λ | 0.003 | 0.193 | |||
| χ2 | 3770.20 | 1094.90 | |||
| df | 60 | 42 | |||
| p | <0.0001 | <0.0001 | |||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacobs, D.S.; Bastian, A. High Duty Cycle Echolocation May Constrain the Evolution of Diversity within Horseshoe Bats (Family: Rhinolophidae). Diversity 2018, 10, 85. https://doi.org/10.3390/d10030085
Jacobs DS, Bastian A. High Duty Cycle Echolocation May Constrain the Evolution of Diversity within Horseshoe Bats (Family: Rhinolophidae). Diversity. 2018; 10(3):85. https://doi.org/10.3390/d10030085
Chicago/Turabian StyleJacobs, David S., and Anna Bastian. 2018. "High Duty Cycle Echolocation May Constrain the Evolution of Diversity within Horseshoe Bats (Family: Rhinolophidae)" Diversity 10, no. 3: 85. https://doi.org/10.3390/d10030085
APA StyleJacobs, D. S., & Bastian, A. (2018). High Duty Cycle Echolocation May Constrain the Evolution of Diversity within Horseshoe Bats (Family: Rhinolophidae). Diversity, 10(3), 85. https://doi.org/10.3390/d10030085