Synergy Effect of Combining Fluorescence and Mid Infrared Fiber Spectroscopy for Kidney Tumor Diagnostics
Abstract
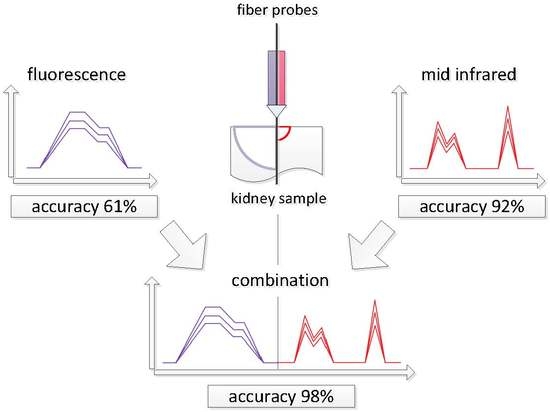
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Spectroscopic Measurements
2.3. Data Analysis
3. Results and Discussion
4. Conclusions and Outlook
Acknowledgments
Author Contributions
Conflicts of Interest
Ethical Statements
Appendix A. Source Data
References
- Nallala, J.; Diebold, M.D.; Gobinet, C.; Bouché, O.; Sockalingum, G.D.; Piot, O.; Manfait, M. Infrared Spectral Histopathology for Cancer Diagnosis: A Novel Approach for Automated Pattern Recognition of Colon Adenocarcinoma. Analyst 2014, 139, 4005–4015. [Google Scholar] [CrossRef] [PubMed]
- Fullwood, L.M.; Clemens, G.; Griffiths, D.; Ashton, K.; Dawson, T.P.; Lea, R.W.; Davis, C.; Bonnier, F.; Byrned, H.J.; Baker, M.J. Investigating the Use of Raman and Immersion Raman Spectroscopy for Spectral Histopathology of Metastatic Brain Cancer and Primary Sites of Origin. Anal. Methods 2014, 6, 3948–3961. [Google Scholar] [CrossRef]
- American Cancer Society. Cancer Facts and Figures 2016; American Cancer Society: Atlanta, GA, USA, 2016. [Google Scholar]
- Matin, J.S.; Pagliaro, L.C.; Wood, C.G.; Tannir, N.M.; Anderson, M.D. Manual of Medical Oncology, 2nd ed.; McGraw-Hill: New York, NY, USA, 2011; pp. 905–924. ISBN 978-0-07-184794-0. [Google Scholar]
- Sachdeva, K.; Harris, J.E. Renal Cell Carcinoma. Available online: http://emedicine.medscape.com/article/281340-overview (accessed on 16 September 2017).
- Yang, H.; He, B.; Dai, X.; Satpathy, M.; Yang, L.; Jiang, H. FMTPen: A Miniaturized Handheld Fluorescence Molecular Tomography Probe for Image-Guided Cancer Surgery. Photonics 2015, 2, 279–287. [Google Scholar] [CrossRef]
- Ozaki, M.; Kagawa, K.; Arimoto, H.; Kominami, Y.; Sanomura, Y.; Yoshida, S.; Seo, M.W.; Kawahito, S.; Tanaka, S. Spectral Study of Metabolism-Based Autofluorescence and White-Light Reflectance for Endoscopic Tumor Imaging. In Proceedings of the 2015 37th Annual International Conference of the IEEE on Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 5879–5883. [Google Scholar]
- Valdes, P.A.; Kim, A.; Leblond, F.; Conde, O.M.; Harris, B.T.; Paulsen, K.D.; Wilson, B.C.; Roberts, D.W. Combined Fluorescence and Reflectance Spectroscopy for in Vivo Quantification of Cancer Biomarkers in Low- and High-Grade Glioma Surgery. J. Biomed. Opt. 2011, 16, 116007. [Google Scholar] [CrossRef] [PubMed]
- Bunaciu, A.A.; Fleschin, Ş.; Aboul-Enein, H.Y. Cancer Diagnosis by FT-IR Spectrophotometry. Rev. Roum. Chim. 2015, 60, 415–426. [Google Scholar]
- Stuart, B.H. Biological Applications of Infrared Spectroscopy, 1st ed.; Ando, D.J., Ed.; Wiley: Hoboken, NJ, USA, 2012; ISBN 978-0471974147. [Google Scholar]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier Transform IR Spectroscopy to Analyze Biological Materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrotra, R.; Tyagi, G.; Jangir, D.K.; Dawar, R.; Gupta, N. Analysis of Ovarian Tumor Pathology by Fourier Transform Infrared Spectroscopy. J. Ovarian Res. 2010, 3, 27. [Google Scholar] [CrossRef] [PubMed]
- Sablinskas, V.; Urboniene, V.; Ceponkus, J.; Laurinavicius, A.; Dasevicius, D.; Jankevicius, F.; Hendrixson, V.; Koch, E.; Steiner, G. Infrared Spectroscopic Imaging of Renal Tumour Tissue. J. Biomed. Opt. 2011, 16, 096006. [Google Scholar] [CrossRef] [PubMed]
- Lima, C.; Goulart, V.; Côrrea, L.; Pereira, T.; Zezell, D. ATR-FTIR Spectroscopy for the Assessment of Biochemical Changes in Skin Due to Cutaneous Squamous Cell Carcinoma. Int. J. Mol. Sci. 2015, 16, 6621–6630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, A.; Khurana, M.; Moriyama, Y.; Wilson, B.C. Quantification of in Vivo Fluorescence Decoupled From the Effects of Tissue Optical Properties Using Fiber-Optic Spectroscopy Measurements. J. Biomed. Opt. 2010, 15, 067006. [Google Scholar] [CrossRef] [PubMed]
- Ollesch, J.; Zaczek, M.; Heise, H.M.; Theisen, O.; Großerüschkamp, F.; Schmidt, R.; Morgenroth, K.; Philippou, S.; Kemen, M.; Gerwert, K. Clinical Application of Infrared Fibre-Optic Probes for the Discrimination of Colorectal Cancer Tissues and Cancer Grades. Vib. Spectrosc. 2017, 91, 99–110. [Google Scholar] [CrossRef]
- Bindig, U.; Müller, G. Fibre-Optic Laser-Assisted Infrared Tumour Diagnostics (FLAIR). J. Phys. D Appl. Phys. 2005, 38, 2716–2731. [Google Scholar] [CrossRef]
- Heise, H.M.; Küpper, L.; Butvina, L.N. Bio-Analytical Applications of Mid-Infrared Spectroscopy Using Silver Halide Fiber-Optic Probes. Spectrochim. Acta Part B 2002, 57, 1649–1663. [Google Scholar] [CrossRef]
- Kang, J.W.; Lue, N.; Kong, C.-R.; Barman, I.; Dingari, N.C.; Goldfless, S.J.; Niles, J.C.; Dasari, R.R.; Feld, M.S. Combined Confocal Raman and Quantitative Phase Microscopy System for Biomedical Diagnosis. Biomed. Opt. Express 2011, 2, 2484–2492. [Google Scholar] [CrossRef] [PubMed]
- Tunnell, J.W.; Desjardins, A.E.; Galindo, L.; Georgakoudi, I.; McGee, S.A.; Mirkovic, J.; Mueller, M.G.; Nazemi, J.; Nguyen, F.T.; Wax, A.; et al. Instrumentation for Multi-Modal Spectroscopic Diagnosis of Epithelial Dysplasia. Technol. Cancer Res. Treat. 2003, 2, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Volynskaya, Z.; Haka, A.S.; Bechtel, K.L.; Fitzmaurice, M.; Shenk, R.; Wang, N.; Nazemi, J.; Dasari, R.R.; Feld, M.S. Diagnosing Breast Cancer Using Diffuse Reflectance Spectroscopy and Intrinsic Fluorescence Spectroscopy. J. Biomed. Opt. 2008, 13, 024012. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.K.; Mirabal, Y.N.; Atkinson, E.N.; Cox, D.; Malpica, A.; Follen, M.; Richards-Kortum, R.J. Combined Reflectance and Fluorescence Spectroscopy for In Vivo Detection of Cervical pre-Cancer. J. Biomed. Opt. 2005, 10, 024031. [Google Scholar] [CrossRef] [PubMed]
- Bogomolov, A.; Zabarylo, U.; Kirsanov, D.; Belikova, V.; Ageev, V.; Usenov, I.; Galyanin, V.; Minet, O.; Sakharova, T.; Danielyan, G.; et al. LED-Based Near Infrared Sensor for Human Kidney Tumor Diagnostics. Sensors 2017, 17, 1914. [Google Scholar] [CrossRef] [PubMed]
- Artyushenko, V.; Schulte, F.; Zabarylo, U.; Berlien, H.-P.; Usenov, I.; Saeb Gilani, T.; Eichler, H.J.; Pieszczek, Ł.; Bogomolov, A.; Krause, H.; et al. LED-Based Near Infrared Sensor for Cancer Diagnostics. Proc. SPIE 2015, 9537, 953720. [Google Scholar] [CrossRef]
- Fuhrman, S.A.; Lasky, L.C.; Limas, C. Prognostic Significance of Morphologic Parameters in Renal Cell Carcinoma. C. Am J. Surg. Pathol. 1982, 6, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Moch, H.; Artibani, W.; Delahunt, B.; Ficarra, V.; Knuechel, R.; Montorsi, F.; Patard, J.-J.; Stief, C.G.; Sulser, T.; Wild, P.J. Reassessing the Current UICC/AJCC TNM Staging for Renal Cell Carcinoma. Eur. Urol. 2009, 56, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Artyushenko, V.; Schulte, F.; Zabarylo, U.; Berlien, H.-P.; Usenov, I.; Saeb Gilani, T.; Eichler, H.; Pieszczek, Ł.; Bogomolov, A.; Krause, H.; et al. Spectral Fiber Sensors for Cancer Diagnostics In Vitro. Proc. SPIE 2015, 9537, 953720. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal Component Analysis. Chemometr. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Brereton, R.G. Chemometrics for Pattern Recognition; John Wiley & Sons, Ltd.: Bristol, UK, 2009; pp. 199–200. ISBN 978-0-470-98725-4. [Google Scholar]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Gajjar, K.; Trevisan, J.; Owens, G.; Keating, P.J.; Wood, N.J.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Martin, F.L. Fourier-Transform Infrared Spectroscopy Coupled with a Classification Machine for The Analysis of Blood Plasma or Serum: A Novel Diagnostic Approach for Ovarian Cancer. Analyst 2013, 138, 3917. [Google Scholar] [CrossRef] [PubMed]
- Krause, H.; Department of Urolgy, Charité Universitätsmedizin, Berlin, Germany. Personal communication.
- Khanmohammadi, M.; Ansari, M.A.; Garmarudi, A.B.; Hassanzadeh, G.; Garoosi, G. Cancer Diagnosis by Discrimination Between Normal and Malignant Human Blood Samples Using Attenuated Total Reflectance-Fourier Transform Infrared Spectroscopy. Cancer Investig. 2007, 25, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.T.; Wong, R.K.; Caputo, T.A.; Godwin, T.A.; Rigas, B. Infrared Spectroscopy of Exfoliated Human Cervical Cells: Evidence of Extensive Structural Changes during Carcinogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 10988–10992. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Thomas, G.J., Jr. Vibrational analysis of nucleic acids. IV. Normal modes of the DNA phosphodiester structure modeled by diethyl phosphate. Biopolymers 1996, 39, 813–835. [Google Scholar] [CrossRef]
- Delahunt, B.; Bethwaite, P.B.; McCredie, M.R.; Nacey, J.N. The Evolution of Collagen Expression in Sarcomatoid Renal Cell Carcinoma. Hum. Pathol. 2007, 38, 1372–1377. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, K.; Galvan, J.; Myakov, A.; Lacy, A.; Lotan, R.; Richards-Kortum, R.J. Realistic Three-Dimensional Epithelial Tissue Phantoms for Biomedical Optics. J. Biomed. Opt. 2002, 7, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Bogomolov, A. Multivariate Process Trajectories: Capture, Resolution and Analysis. Chemometr. Intell. Lab. Syst. 2011, 108, 49–63. [Google Scholar] [CrossRef]
- De Groot, P.G.; Swierenga, H.; Postma, G.J.; Melssen, W.J.; Buydens, L.M.C. Effect on the PLS Prediction of Yarn Properties Combining Raman and Infrared Measurements and Applying Wavelength Selection. Appl. Spectrosc. 2003, 57, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Bogomolov, A.; Engler, M.; Melichar, M.; Wigmore. In-Line Analysis of a Fluid Bed Pellet Coating Process Using a Combination of Near Infrared and Raman Spectroscopy. J. Chemometr. 2010, 24, 544–557. [Google Scholar] [CrossRef]
- Heberger, K. Sum of ranking differences compares methods or models fairly. Trends TRAC Anal. Chem. 2010, 29, 101–109. [Google Scholar] [CrossRef]
- Kollar-Hunek, K.; Heberger, K. Method and model comparison by sum of ranking differences in cases of repeated observations (ties). Chemometr. Intell. Lab. Syst. 2013, 127, 139–146. [Google Scholar] [CrossRef]
- Heberger, K.; Kollar-Hunek, K. Sum of ranking differences for method discrimination and its validation: Comparison of ranks with random numbers. J. Chemometr. 2011, 25, 151–158. [Google Scholar] [CrossRef]




| Method | Preprocessing | TP | FP | TN | FN | %Ac | %Sn | %Sp |
|---|---|---|---|---|---|---|---|---|
| Calibration 1 | ||||||||
| Fluorescence | none | 37 | 20 | 21 | 14 | 63 | 73 | 51 |
| SNV 2 | 32 | 12 | 29 | 19 | 66 | 63 | 71 | |
| MIR | none | 38 | 1 | 40 | 13 | 85 | 75 | 98 |
| SNV | 42 | 2 | 39 | 9 | 88 | 82 | 95 | |
| 2D 3 | 45 | 4 | 37 | 6 | 89 | 88 | 90 | |
| 2D + SNV | 49 | 0 | 41 | 2 | 98 | 96 | 100 | |
| Fluorescence | MIR | AS 4 | AS | 39 | 13 | 28 | 12 | 73 | 76 | 68 |
| AS | 2D + AS | 44 | 8 | 33 | 7 | 84 | 86 | 80 | |
| SNV | SNV | 48 | 0 | 41 | 3 | 97 | 94 | 100 | |
| SNV | 2D + SNV | 51 | 0 | 41 | 0 | 100 | 100 | 100 | |
| Cross-validation 5 | ||||||||
| Fluorescence | none | 32 | 22 | 19 | 19 | 55 | 63 | 46 |
| SNV | 27 | 12 | 29 | 24 | 61 | 53 | 71 | |
| MIR | none | 38 | 3 | 38 | 13 | 83 | 75 | 93 |
| SNV | 42 | 4 | 37 | 9 | 86 | 82 | 90 | |
| 2D | 45 | 5 | 36 | 6 | 88 | 88 | 88 | |
| 2D + SNV | 45 | 1 | 40 | 6 | 92 | 88 | 98 | |
| Fluorescence | MIR | AS | AS | 35 | 15 | 26 | 16 | 66 | 69 | 63 |
| AS | 2D + AS | 37 | 9 | 32 | 14 | 75 | 73 | 78 | |
| SNV | SNV | 47 | 0 | 41 | 4 | 96 | 92 | 100 | |
| SNV | 2D + SNV | 49 | 0 | 41 | 2 | 98 | 96 | 100 | |
| Random-subset validation 6 | ||||||||
| Fluorescence | none | 61 | 70 | 50 | ||||
| SNV | 65 | 61 | 70 | |||||
| MIR | none | 84 | 75 | 95 | ||||
| SNV | 88 | 83 | 94 | |||||
| 2D | 89 | 87 | 91 | |||||
| 2D + SNV | 95 | 92 | 99 | |||||
| Fluorescence | MIR | AS | AS | 71 | 75 | 66 | ||||
| AS | 2D + AS | 81 | 83 | 80 | |||||
| SNV | SNV | 96 | 94 | 100 | |||||
| SNV | 2D + SNV | 99 | 98 | 100 | |||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bogomolov, A.; Belikova, V.; Zabarylo, U.J.; Bibikova, O.; Usenov, I.; Sakharova, T.; Krause, H.; Minet, O.; Feliksberger, E.; Artyushenko, V. Synergy Effect of Combining Fluorescence and Mid Infrared Fiber Spectroscopy for Kidney Tumor Diagnostics. Sensors 2017, 17, 2548. https://doi.org/10.3390/s17112548
Bogomolov A, Belikova V, Zabarylo UJ, Bibikova O, Usenov I, Sakharova T, Krause H, Minet O, Feliksberger E, Artyushenko V. Synergy Effect of Combining Fluorescence and Mid Infrared Fiber Spectroscopy for Kidney Tumor Diagnostics. Sensors. 2017; 17(11):2548. https://doi.org/10.3390/s17112548
Chicago/Turabian StyleBogomolov, Andrey, Valeria Belikova, Urszula J. Zabarylo, Olga Bibikova, Iskander Usenov, Tatiana Sakharova, Hans Krause, Olaf Minet, Elena Feliksberger, and Viacheslav Artyushenko. 2017. "Synergy Effect of Combining Fluorescence and Mid Infrared Fiber Spectroscopy for Kidney Tumor Diagnostics" Sensors 17, no. 11: 2548. https://doi.org/10.3390/s17112548
APA StyleBogomolov, A., Belikova, V., Zabarylo, U. J., Bibikova, O., Usenov, I., Sakharova, T., Krause, H., Minet, O., Feliksberger, E., & Artyushenko, V. (2017). Synergy Effect of Combining Fluorescence and Mid Infrared Fiber Spectroscopy for Kidney Tumor Diagnostics. Sensors, 17(11), 2548. https://doi.org/10.3390/s17112548