Porous Silicon Based Resonant Mirrors for Biochemical Sensing
Abstract
:1. Introduction
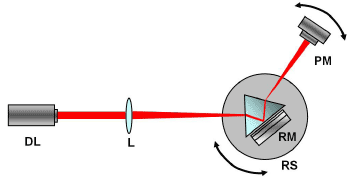
2. Experimental
3. Results and Discussion
4. Conclusions
Acknowledgments
References and Notes
- Lechuga, L.M.; Tamayo, J.; Álvarez, M.; Carrascosa, L.G.; Yufera, A.; Doldán, R.; Peralías, E.; Rueda, A.; Plaza, A.J.; Zinoviev, K.; Domínguez, C.; Zaballos, A.; Moreno, M.; Martínez-A, C.; Wenn, D.; Harris, N.; Bringer, C.; Bardinal, V.; Camps, T.; Vergenègre, C.; Fontaine, C.; Díaz, V.; Bernad, A. A highly sensitive microsystem based on nanomechanical biosensors for genomics applications. Sensor. Actuat. B-Chem. 2006, 118, 2–10. [Google Scholar]
- Pacholski, C.; Sartor, M.; Sailor, M. J.; Cunin, F.; Miskelly, G. M. Biosensing using porous silicon double-layer interferometers: Reflective interferometric fourier transform spectroscopy. J. Am. Chem. Soc. 2005, 127, 11636–11645. [Google Scholar]
- De Stefano, L.; Rotiroti, L.; Rea, I.; Rendina, I.; Moretti, L.; Di Francia, G.; Massera, E.; Lamberti, A.; Arcari, P.; Sangez, C. Porous silicon-based optical biochips. J. Opt. A-Pure Appl. Op. 2006, 8, S540–S544. [Google Scholar]
- Lee, M.R.; Fauchet, P.M. Two-dimensional silicon photonic crystal based biosensing platform for protein detection. Opt. Express 2007, 15, 4530–4535. [Google Scholar]
- Cush, R.; Cronin, J.M.; Stewart, W.J.; Maule, C.H.; Molloy, J.; Goddard, N.J. The resonant mirror: a novel optical biosensor for direct sensing of biomolecular interactions. I: Principle of operation and associated instrumentation. Biosens Bioelectron. 1993, 8, 347–353. [Google Scholar]
- Baldini, F.; Chester, A.N.; Homola, J.; Martellucci, S. Optical Chemical Sensors.; Springer: Dordrecht, The Netherlands, 2006. [Google Scholar]
- Chapron, J.; Alekseev, S.A.; Lysenko, V.; Zaitsev, V.N.; Barbier, D. Analysis of interaction between chemical agents and porous Si nanostructures using optical sensing properties of infra-red Rugate filters. Sensor. Actuat. B-Chem. 2007, 120, 706–711. [Google Scholar]
- Ouyang, H.; Chrtistophersen, M.; Viard, R.; Miller, B.L.; Fauchet, P.M. Macroporous silicon microcavities for macromolecule detection. Adv. Funct. Mater. 2005, 15, 1851–1859. [Google Scholar]
- Saarinen, J.J.; Weiss, S.M.; Fauchet, P.M.; Sipe, J.E. Optical sensor based on resonant porous silicon structures. Opt. Express 2005, 13, 3754–3764. [Google Scholar]
- Khardani, M.; Bouaïcha, M.; Bessaïs, B. Bruggeman effective medium approach for modelling optical properties of porous silicon: comparison with experiment. Phys. Stat. Sol. 2007, 4, 1986–1990. [Google Scholar]
- Pirasteh, P.; Charrier, J.; Dumeige, Y.; Haesaert, S.; Joubert, P. Optical loss of porous silicon and oxidized porous silicon planar waveguides. J. Appl. Phys. 2007, 101, 083110. [Google Scholar]
- Ulrich, R.; Torge, R. Measurement of thin film parameters with a prism coupler. Appl. Optics 1973, 12, 2901–2908. [Google Scholar]
- Rong, G.; Najmaie, A.; Sipe, J.E.; Weiss, S. M. Nanoscale porous silicon waveguide for label-free DNA sensing. Biosens. Bioelectron. 2008, 23, 1572–1576. [Google Scholar]
- Descrovi, E.; Frascella, F.; Sciacca, B.; Geobaldo, F.; Dominici, L.; Michelotti, F. Coupling of surface waves in highly defined one-dimensional porous silicon photonic crystals for gas sensing applications. Appl. Phys. Lett. 2007, 91, 241109. [Google Scholar]






© 2008 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
De Tommasi, E.; De Stefano, L.; Rea, I.; Di Sarno, V.; Rotiroti, L.; Arcari, P.; Lamberti, A.; Sanges, C.; Rendina, I. Porous Silicon Based Resonant Mirrors for Biochemical Sensing. Sensors 2008, 8, 6549-6556. https://doi.org/10.3390/s8106549
De Tommasi E, De Stefano L, Rea I, Di Sarno V, Rotiroti L, Arcari P, Lamberti A, Sanges C, Rendina I. Porous Silicon Based Resonant Mirrors for Biochemical Sensing. Sensors. 2008; 8(10):6549-6556. https://doi.org/10.3390/s8106549
Chicago/Turabian StyleDe Tommasi, Edoardo, Luca De Stefano, Ilaria Rea, Valentina Di Sarno, Lucia Rotiroti, Paolo Arcari, Annalisa Lamberti, Carmen Sanges, and Ivo Rendina. 2008. "Porous Silicon Based Resonant Mirrors for Biochemical Sensing" Sensors 8, no. 10: 6549-6556. https://doi.org/10.3390/s8106549
APA StyleDe Tommasi, E., De Stefano, L., Rea, I., Di Sarno, V., Rotiroti, L., Arcari, P., Lamberti, A., Sanges, C., & Rendina, I. (2008). Porous Silicon Based Resonant Mirrors for Biochemical Sensing. Sensors, 8(10), 6549-6556. https://doi.org/10.3390/s8106549