Screening of Bacterial Quorum Sensing Inhibitors in a Vibrio fischeri LuxR-Based Synthetic Fluorescent E. coli Biosensor
Abstract
:1. Introduction
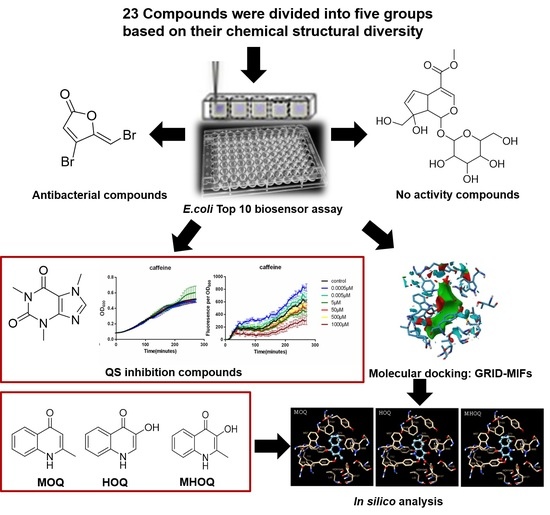
2. Results
2.1. Screening a Panel of Potential Quorum Sensing Inhibitors
2.2. Computation of GRID-MIFs
2.3. Docking and Interaction of Selected Compounds
3. Discussion
4. Materials and Methods
4.1. Library of Tested Chemical Compounds
4.2. Bacterial Strains
4.3. Growth Media and Glycerol Stocks Preparation
4.4. E. coli Top10 Fluorescent Biosensor Assay
4.5. Protein Structure File, Ligand Database
4.6. Molecular Docking Studies
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum Sensing in Bacteria: The LuxR-LuxI Family of Cell Density-Responsive Transcriptional Regulatorst. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutherford, S.T.; Bassler, B. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- Bassler, B.; Losick, R. Bacterially Speaking. Cell 2006, 125, 237–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passador, L.; Cook, J.; Gambello, M.; Rust, L.; Iglewski, B. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 1993, 260, 1127–1130. [Google Scholar] [CrossRef]
- Ochsner, U.A.; Reiser, J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 6424–6428. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.K.; Schaefer, A.L.; Parsek, M.R.; Moninger, T.O.; Welsh, M.J.; Greenberg, E.P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 2000, 407, 762–764. [Google Scholar] [CrossRef]
- Vannini, A.; Volpari, C.; Gargioli, C.; Muraglia, E.; Cortese, R.; De Francesco, R.; Neddermann, P.; Di Marco, S. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002, 21, 4393–4401. [Google Scholar] [CrossRef]
- Hanzelka, B.L.; Greenberg, E.P. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 1995, 177, 815–817. [Google Scholar] [CrossRef] [Green Version]
- Stevens, A.M.; Dolan, K.M.; Greenberg, E.P. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. USA 1994, 91, 12619–12623. [Google Scholar] [CrossRef] [Green Version]
- Chai, Y.; Winans, S.C. Site-directed mutagenesis of a LuxR-type quorum-sensing transcription factor: Alteration of autoinducer specificity. Mol. Microbiol. 2004, 51, 765–776. [Google Scholar] [CrossRef]
- Choi, S.H.; Greenberg, E.P. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc. Natl. Acad. Sci. USA 1991, 88, 11115–11119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiratisin, P.; Tucker, K.D.; Passador, L. LasR, a Transcriptional Activator of Pseudomonas aeruginosa Virulence Genes, Functions as a Multimer. J. Bacteriol. 2002, 184, 4912–4919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamb, J.R.; Patel, H.; Montminy, T.; Wagner, V.E.; Iglewski, B.H. FunctionalDomains of the RhlR Transcriptional Regulator ofPseudomonasaeruginosa. J. Bacteriol. 2003, 185, 7129–7139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Z.-Q.; Smyth, A.J.; Gao, P.; Qin, Y.; Farrand, S.K. Mutational Analysis of TraR. J. Boil. Chem. 2003, 278, 13173–13182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shadel, G.S.; Young, R.F.; Baldwin, T.O. Use of regulated cell lysis in a lethal genetic selection in Escherichia coli: Identification of the autoinducer-binding region of the LuxR protein from Vibrio fischeri ATCC 7744. J. Bacteriol. 1990, 172, 3980–3987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slock, J.; VanRiet, D.; Kolibachuk, D.; Greenberg, E.P. Critical regions of the Vibrio fischeri luxR protein defined by mutational analysis. J. Bacteriol. 1990, 172, 3974–3979. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.-G.; Pappas, K.M.; Brace, J.L.; Miller, P.C.; Oulmassov, T.; Molyneaux, J.M.; Anderson, J.C.; Bashkin, J.K.; Winans, S.C.; Joachimiak, A. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 2002, 417, 971–974. [Google Scholar] [CrossRef]
- Müh, U.; Hare, B.J.; Duerkop, B.A.; Schuster, M.; Hanzelka, B.L.; Heim, R.; Olson, E.R.; Greenberg, E.P. A structurally unrelated mimic of a Pseudomonas aeruginosa acyl-homoserine lactone quorum-sensing signal. Proc. Natl. Acad. Sci. USA 2006, 103, 16948–16952. [Google Scholar] [CrossRef] [Green Version]
- Koch, B.; Liljefors, T.; Persson, T.; Nielsen, J.; Kjelleberg, S.; Givskov, M. The LuxR receptor: The sites of interaction with quorum-sensing signals and inhibitors. Microbiology 2005, 151, 3589–3602. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Yin, B.; Qian, L.; Zeng, Z.; Yang, Z.; Li, H.; Lu, Y.; Zhou, S. Screening for novel quorum-sensing inhibitors to interfere with the formation of Pseudomonas aeruginosa biofilm. J. Med. Microbiol. 2011, 60, 1827–1834. [Google Scholar] [CrossRef]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2015, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.A.; Hoven, A.D.; Cook, A.M. Therapeutic frontiers: Preventing and treating infectious diseases by inhibiting bacterial quorum sensing. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 27, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Kociolek, M. Quorum-Sensing Inhibitors and Biofilms. Anti Infect. Agents Med. Chem. 2009, 8, 315–326. [Google Scholar] [CrossRef]
- Choudhary, S.; Schmidt-Dannert, C. Applications of quorum sensing in biotechnology. Appl. Microbiol. Biotechnol. 2010, 86, 1267–1279. [Google Scholar] [CrossRef]
- Clatworthy, A.E.; Pierson, E.; Hung, D.T. Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Methods 2007, 3, 541–548. [Google Scholar] [CrossRef]
- Kalia, V.C. Quorum sensing inhibitors: An overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef]
- Chen, F.; Gao, Y.; Chen, X.; Yu, Z.; Li, X. Quorum Quenching Enzymes and Their Application in Degrading Signal Molecules to Block Quorum Sensing-Dependent Infection. Int. J. Mol. Sci. 2013, 14, 17477–17500. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, P.; Zhang, G.; Xu, L.; Wang, D.; Wang, L.; Zeng, X.; Wang, Y. Design, synthesis and antibacterial activity of novel andrographolide derivatives. Bioorg. Med. Chem. 2010, 18, 4269–4274. [Google Scholar] [CrossRef]
- Qin, X.; Thota, G.K.; Singh, R.; Balamurugan, R.; Goycoolea, F.M. Synthetic homoserine lactone analogues as antagonists of bacterial quorum sensing. Bioorg. Chem. 2020, 98, 103698. [Google Scholar] [CrossRef]
- Yang, L.; Rybtke, M.T.; Jakobsen, T.H.; Hentzer, M.; Bjarnsholt, T.; Givskov, M.; Tolker-Nielsen, T. Computer-Aided Identification of Recognised Drugs as Pseudomonas aeruginosa Quorum-Sensing Inhibitors. Antimicrob. Agents Chemother. 2009, 53, 2432–2443. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.; Qian, L.; Cao, L.; Tan, H.; Huang, Y.; Xue, X.; Shen, Y.; Zhou, S. Virtual screening for novel quorum sensing inhibitors to eradicate biofilm formation of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2008, 79, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Annapoorani, A.; Umamageswaran, V.; Parameswari, R.; Pandian, S.K.; Ravi, A.V. Computational discovery of putative quorum sensing inhibitors against LasR and RhlR receptor proteins of Pseudomonas aeruginosa. J. Comput. Mol. Des. 2012, 26, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Y.-Y.; Chua, S.L.; Chen, Y.; Rice, S.A.; Kjelleberg, S.; Nielsen, T.E.; Yang, L.; Givskov, M. Identification of Five Structurally Unrelated Quorum-Sensing Inhibitors of Pseudomonas aeruginosa from a Natural-Derivative Database. Antimicrob. Agents Chemother. 2013, 57, 5629–5641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodford, P.J. A Computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 1985, 28, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Carosati, E.; Sciabola, S.; Cruciani, G. Hydrogen Bonding Interactions of Covalently Bonded Fluorine Atoms: From Crystallographic Data to a New Angular Function in the GRID Force Field. J. Med. Chem. 2004, 47, 5114–5125. [Google Scholar] [CrossRef] [PubMed]
- Ahlström, M.M.; Ridderström, M.; Luthman, A.K.; Zamora, I. Virtual Screening and Scaffold Hopping Based on GRID Molecular Interaction Fields. J. Chem. Inf. Model. 2005, 45, 1313–1323. [Google Scholar] [CrossRef]
- Sciabola, S.; Stanton, R.V.; Mills, J.E.; Flocco, M.M.; Baroni, M.; Cruciani, G.; Perruccio, F.; Mason, J.S. High-Throughput Virtual Screening of Proteins Using GRID Molecular Interaction Fields. J. Chem. Inf. Model. 2009, 50, 155–169. [Google Scholar] [CrossRef]
- Sanjurjo, C.V.; Engwer, C.; Qin, X.; Hembach, L.; Verdía-Cotelo, T.; Remuñán-López, C.; Vila-Sanjurjo, A.; Goycoolea, F.M. A single intracellular protein governs the critical transition from an individual to a coordinated population response during quorum sensing: Origins of primordial language. bioRxiv 2016, 074369. [Google Scholar] [CrossRef] [Green Version]
- Skandamis, P.N.; Nychas, G.-J. Quorum Sensing in the Context of Food Microbiology. Appl. Environ. Microbiol. 2012, 78, 5473–5482. [Google Scholar] [CrossRef] [Green Version]
- Galloway, W.R.; Hodgkinson, J.T.; Bowden, S.; Welch, M.; Spring, D.R. Applications of small molecule activators and inhibitors of quorum sensing in Gram-negative bacteria. Trends Microbiol. 2012, 20, 449–458. [Google Scholar] [CrossRef]
- Takano, E. γ-Butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 2006, 9, 287–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gram, L.; De Nys, R.; Maximilien, R.; Givskov, M.; Steinberg, P.; Kjelleberg, S. Inhibitory Effects of Secondary Metabolites from the Red Alga Delisea pulchra on Swarming Motility of Proteus mirabilis. Appl. Environ. Microbiol. 1996, 62, 4284–4287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Givskov, M.; De Nys, R.; Manefield, M.; Gram, L.; Maximilien, R.; Eberl, L.; Molin, S.; Steinberg, P.; Kjelleberg, S. Eukaryotic interference with homoserine lactone-mediated prokaryotuc signalling. J. Bacteriol. 1996, 178, 6618–6622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manefield, M.; De Nys, R.; Naresh, K.; Roger, R.; Givskov, M.; Peter, S.; Kjelleberg, S. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 1999, 145, 283–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, D.; Sims, J.J.; Wood, T.K. Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ. Microbiol. 2001, 3, 731–736. [Google Scholar] [CrossRef]
- Defoirdt, T.; Miyamoto, C.M.; Wood, T.K.; Meighen, E.A.; Sorgeloos, P.; Verstraete, W.; Bossier, P. The natural furanone (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone disrupts quorum sensing-regulated gene expression in Vibrio harveyi by decreasing the DNA-binding activity of the transcriptional regulator protein luxR. Environ. Microbiol. 2007, 9, 2486–2495. [Google Scholar] [CrossRef] [Green Version]
- Lönn-Stensrud, J.; Landin, M.A.; Benneche, T.; Petersen, F.C.; Scheie, A.A. Furanones, potential agents for preventing Staphylococcus epidermidis biofilm infections? J. Antimicrob. Chemother. 2009, 63, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef]
- Manefield, M.; Rasmussen, T.B.; Henzter, M.; Andersen, J.B.; Steinberg, P.; Kjelleberg, S.; Givskov, M. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 2002, 148, 1119–1127. [Google Scholar] [CrossRef] [Green Version]
- Steenackers, H.P.; Levin, J.; Janssens, J.C.; De Weerdt, A.; Balzarini, J.; Vanderleyden, J.; De Vos, D.E.; De Keersmaecker, S.C.J. Structure–activity relationship of brominated 3-alkyl-5-methylene-2(5H)-furanones and alkylmaleic anhydrides as inhibitors of Salmonella biofilm formation and quorum sensing regulated bioluminescence in Vibrio harveyi. Bioorganic Med. Chem. 2010, 18, 5224–5233. [Google Scholar] [CrossRef]
- Janssens, J.C.A.; Steenackers, H.; Robijns, S.; Gellens, E.; Levin, J.; Zhao, H.; Hermans, K.; De Coster, D.; Verhoeven, T.L.; Marchal, K.; et al. Brominated Furanones Inhibit Biofilm Formation by Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 2008, 74, 6639–6648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponnusamy, K.; Paul, D.; Kweon, J.H. Inhibition of Quorum Sensing Mechanism andAeromonas hydrophilaBiofilm Formation by Vanillin. Environ. Eng. Sci. 2009, 26, 1359–1363. [Google Scholar] [CrossRef]
- Niu, C.; Afre, S.; Gilbert, E.S. Subinhibitory concentrations of cinnamaldehyde interfere with quorum sensing. Lett. Appl. Microbiol. 2006, 43, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Brackman, G.; Defoirdt, T.; Miyamoto, C.; Bossier, P.; Van Calenbergh, S.; Nelis, H.J.; Coenye, T. Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in Vibrio spp. by decreasing the DNA-binding activity of the quorum sensing response regulator LuxR. BMC Microbiol. 2008, 8, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.-Y.; Krishnan, T.; Wang, H.; Chen, Y.; Yin, W.-F.; Chong, Y.-M.; Tan, L.Y.; Chong, T.M.; Chan, K.-G. Non-antibiotic quorum sensing inhibitors acting against N-acyl homoserine lactone synthase as druggable target. Sci. Rep. 2014, 4, 7245. [Google Scholar] [CrossRef] [PubMed]
- Furuya, T.; Arai, Y.; Kino, K. Biotechnological Production of Caffeic Acid by Bacterial Cytochrome P450 CYP199A2. Appl. Environ. Microbiol. 2012, 78, 6087–6094. [Google Scholar] [CrossRef] [Green Version]
- Hayman, M.; Kam, P.C. Capsaicin: A review of its pharmacology and clinical applications. Curr. Anaesth. Crit. Care 2008, 19, 338–343. [Google Scholar] [CrossRef]
- Jones, N.L.; Shabib, S.; Sherman, P.M. Capsaicin as an inhibitor of the growth of the gastric pathogen Helicobacter pylori. FEMS Microbiol. Lett. 1997, 146, 223–227. [Google Scholar] [CrossRef]
- Zeyrek, F.Y.; Oguz, E. In vitro activity of capsaicin against Helicobacter pylori. Ann. Microbiol. 2005, 55, 125–127. [Google Scholar]
- Lee, H.S.; Lee, S.Y.; Park, S.H.; Lee, J.H.; Ahn, S.K.; Choi, Y.M.; Choi, D.J.; Chang, J.-H. Antimicrobial medical sutures with caffeic acid phenethyl ester and their in vitro/in vivo biological assessment. MedChemComm 2013, 4, 777. [Google Scholar] [CrossRef]
- Murtaza, G.; Karim, S.; Akram, M.R.; Khan, S.A.; Azhar, S.; Mumtaz, A.; Bin Asad, M.H.H. Caffeic Acid Phenethyl Ester and Therapeutic Potentials. BioMed Res. Int. 2014, 2014, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norizan, S.N.M.; Yin, W.-F.; Chan, K.-G. Caffeine as a Potential Quorum Sensing Inhibitor. Sensors 2013, 13, 5117–5129. [Google Scholar] [CrossRef]
- Koo, H.-J.; Song, Y.S.; Kim, H.-J.; Lee, Y.-H.; Hong, S.-M.; Lim, S.-J.; Kim, B.-C.; Jin, C.; Lim, C.-J.; Park, E.-H. Anti-inflammatory effects of genipin, an active principle of gardenia. Eur. J. Pharmacol. 2004, 495, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-C.; Kim, H.-G.; Lee, S.-A.; Lim, S.; Park, E.-H.; Kim, S.-J.; Lim, C.-J. Genipin-induced apoptosis in hepatoma cells is mediated by reactive oxygen species/c-Jun NH2-terminal kinase-dependent activation of mitochondrial pathway. Biochem. Pharmacol. 2005, 70, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Sanjurjo, C.V.; David, L.; Remuñán-López, C.; Goycoolea, F.M.; Vila-Sanjurjo, A. Effect of the ultrastructure of chitosan nanoparticles in colloidal stability, quorum quenching and antibacterial activities. J. Colloid Interface Sci. 2019, 556, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-H.; Tsai, S.-C.; Lai, C.-H.; Lee, C.-H.; He, Z.S.; Tseng, G.-C. Genipin-cross-linked fucose–chitosan/heparin nanoparticles for the eradication of Helicobacter pylori. Biomaterials 2013, 34, 4466–4479. [Google Scholar] [CrossRef]
- Hwang, Y.-J.; Larsen, J.; Krasieva, T.B.; Lyubovitsky, J.G. Effect of Genipin Crosslinking on the Optical Spectral Properties and Structures of Collagen Hydrogels. ACS Appl. Mater. Interfaces 2011, 3, 2579–2584. [Google Scholar] [CrossRef] [Green Version]
- Canton, B.; Labno, A.; Endy, D. Refinement and standardisation of synthetic biological parts and devices. Nat. Biotechnol. 2008, 26, 787–793. [Google Scholar] [CrossRef]
- Pe’delacq, J.-D.; Cabantous, S.; Tran, T.; Terwilliger, T.C.; Waldo, G.S. Corrigendum: Engineering and characterisation of a superfolder green fluorescent protein. Nat. Biotechnol. 2006, 24, 1170. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.-S.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas Quinolone Signal (PQS): Not Just for Quorum Sensing Anymore. Front. Microbiol. 2018, 8. [Google Scholar] [CrossRef]
- Price-Whelan, A.; Dietrich, L.E.; Newman, D.K. Rethinking ’secondary’ metabolism: Physiological roles for phenazine antibiotics. Nat. Methods 2006, 2, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Pesci, E.C.; Milbank, J.B.J.; Pearson, J.P.; McKnight, S.; Kende, A.S.; Greenberg, E.P.; Iglewski, B.H. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1999, 96, 11229–11234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKnight, S.L.; Iglewski, B.H.; Pesci, E.C. The Pseudomonas Quinolone Signal Regulates rhl Quorum Sensing in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 2702–2708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Moustafa, D.A.; Stergioula, V.; Smith, C.D.; Goldberg, J.B.; Bassler, B. The PqsE and RhlR proteins are an autoinducer synthase–receptor pair that control virulence and biofilm development in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2018, 115, E9411–E9418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahajan-Miklos, S.; Tan, M.; Rahme, L.G.; Ausubel, F.M. Molecular Mechanisms of Bacterial Virulence Elucidated Using a Pseudomonas aeruginosa– Caenorhabditis elegans Pathogenesis Model. Cell 1999, 96, 47–56. [Google Scholar] [CrossRef] [Green Version]
- Dietrich, L.E.; Price-Whelan, A.; Petersen, A.; Whiteley, M.; Newman, D.K. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 2006, 61, 1308–1321. [Google Scholar] [CrossRef]
- Seo, S.; Gao, Y.; Kim, N.; Szubin, R.; Yang, J.; Cho, B.-K.; Palsson, B.O. Revealing genome-scale transcriptional regulatory landscape of OmpR highlights its expanded regulatory roles under osmotic stress in Escherichia coli K-12 MG1655. Sci. Rep. 2017, 7, 2181. [Google Scholar] [CrossRef] [Green Version]
- Jagmann, N.; Brachvogel, H.-P.; Philipp, B. Parastic growth of Pseudomonas aeruginosa in co-culture with the chitinolytic bacterium Aeromonas hydrophila. Environ. Microbiol. 2010, 12, 1787–1802. [Google Scholar] [CrossRef]
- Bou-Abdallah, F.; Chasteen, N.D.; Lesser, M.P. Quenching of superoxide radicals by green fluorecent protein. Biochim. Biophys. Acta 2006, 1760, 1960–1965. [Google Scholar]
- Mavrodi, D.V.; Bonsall, R.F.; Delaney, S.M.; Soule, M.J.; Phillips, G.; Thomashow, L.S. Functional Analysis of Genes for Biosynthesis of Pyocyanin and Phenazine-1-Carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 2001, 183, 6454–6465. [Google Scholar] [CrossRef] [Green Version]
- Haynes, W.C.; Stodola, F.H.; Locke, J.M.; Pridham, T.G.; Conway, H.F.; Sohns, V.E.; Jackson, R.W. PSEUDOMONAS AUREOFACIENS KLUYVER AND PHENAZINE α-CARBOXYLIC ACID, ITS CHARACTERISTIC PIGMENT. J. Bacteriol. 1956, 72, 412–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzola, M.; Cook, R.J.; Thomashow, L.S.; Weller, D.M.; Pierson, L.S. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 1992, 58, 2616–2624. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wilks, J.C.; Danhorn, T.; Ramos, I.; Croal, L.; Newman, D.K. Phenazine-1-Carboxylic Acid Promotes Bacterial Biofilm Development via Ferrous Iron Acquisition. J. Bacteriol. 2011, 193, 3606–3617. [Google Scholar] [CrossRef] [Green Version]
- Pierson, L.S.; Pierson, E.A. Metabolism and function of phenazines in bacteria: Impacts on the behavior of bacteria in the environment and biotechnological processes. Appl. Microbiol. Biotechnol. 2010, 86, 1659–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassett, D.J.; Ma, J.-F.; Elkins, J.G.; McDermott, T.R.; Ochsner, U.A.; West, S.E.H.; Huang, C.-T.; Fredericks, J.; Burnett, S.; Stewart, P.S.; et al. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 1999, 34, 1082–1093. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.K.; Grahl, N.; Okegbe, C.; Dietrich, L.E.; Jacobs, N.J.; Hogan, D.A. Control of Candida albicans Metabolism and Biofilm Formation by Pseudomonas aeruginosa Phenazines. mBio 2013, 4, 00526-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skindersoe, M.E.; Alhede, M.; Phipps, R.; Yang, L.; Jensen, P.Ø.; Rasmussen, T.B.; Bjarnsholt, T.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M. Effects of Antibiotics on Quorum Sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008, 52, 3648–3663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schertzer, J.W.; Boulette, M.L.; Whiteley, M. More than a signal: Non-signaling properties of quorum sensing molecules. Trends Microbiol. 2009, 17, 189–195. [Google Scholar] [CrossRef]
- Kubo, I.; Fujita, K.-I.; Lee, S.H.; Ha, T.J. Antibacterial activity of polygodial. Phytother. Res. 2005, 19, 1013–1017. [Google Scholar] [CrossRef]
- Wang, X.; Yao, X.; Zhu, Z.-A.; Tang, T.; Dai, K.; Sadovskaya, I.; Flahaut, S.; Jabbouri, S. Effect of berberine on Staphylococcus epidermidis biofilm formation. Int. J. Antimicrob. Agents 2009, 34, 60–66. [Google Scholar] [CrossRef]
- Sun, T.; Li, X.-D.; Hong, J.; Liu, C.; Zhang, X.-L.; Zheng, J.-P.; Xu, Y.-J.; Ou, Z.-Y.; Zheng, J.-L.; Yu, D.-J. Inhibitory Effect of Two Traditional Chinese Medicine Monomers, Berberine and Matrine, on the Quorum Sensing System of Antimicrobial-Resistant Escherichia coli. Front. Microbiol. 2019, 10, 2584. [Google Scholar] [CrossRef] [PubMed]
- Aswathanarayan, J.B.; Vittal, R.R. Inhibition of biofilm formation and quorum sensing mediated phenotypes by berberine in Pseudomonas aeruginosa and Salmonella typhimurium. RSC Adv. 2018, 8, 36133–36141. [Google Scholar] [CrossRef] [Green Version]
- Eiden, F.; Wendt, R.; Fenner, H. ChemInform Abstract: PYRONES AND PYRIDONES, PART 74. QUINOLYLIDENE DERIVATIVES. Chem. Informationsdienst 1978, 9, 561–568. [Google Scholar] [CrossRef]
- Evans, D.; Eastwood, F. Synthesis of an arylhydroxytetronimide and of 3-Hydroxy-4(1H)-quinolone derivatives. Aust. J. Chem. 1974, 27, 537. [Google Scholar] [CrossRef]
- Cornforth, J.W.; James, A.T. Structure of a naturally occurring antagonist of dihydrostreptomycin. Biochem. J. 1956, 63, 124–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Compound | Hydrogen Bonding Interactions | Dock Score (Kcal/mol) | Hydrophobic Interactions |
|---|---|---|---|
| OOHL | 4 (including Trp 57) | −7.58 | Leu 40, Tyr 53, Gln 58, Trp 85, Phe101, Ala 105, Ile 110. |
| MOQ (15) | 1 (including Trp 57) | −6.16 | Tyr 53, Val 72, Trp 85, Phe 101, Tyr 102, Ala 105, Met 127 |
| HOQ (16) | 0 | −5.72 | Trp 57, Tyr 61, Asp 70, Val 72, Trp 85, Phe 101, Met 127 |
| MHOQ (17) | 1 (including Trp 57) | −5.83 | Tyr 53, Asp 70, Val 72, Trp 85, Phe 101, Tyr 102, Ala 105, Met 127, Thr 129 |
| PCA (19) | 3 (including Trp 57) | −8.01 | Leu 40, Tyr 53, Tyr 61, Val 72, Trp 85, Phe101, Tyr 102, Ala 105, Met 127 |
| PQS (14) (conf A) | 2 | −8.04 | Leu 40, Tyr 53, Tyr 61, Val 72, Val 73, Trp 85, Phe 101, Tyr 102, Ala 105, Met127, Thr 129 |
| PQS (14) (conf B) | 1 (including Trp 57) | −6.59 | Ala 38, Leu 40, Ala 49, Thr 51, Gln 58, Tyr 61, Phe 62, Val 72, Trp 85, Phe 101, Met127 |
| Number | Compounds | Group 1 | Solvent/Method | Supplier |
|---|---|---|---|---|
| 1 | γ-Valerolactone | Group (1) Lactone analogues | Water | Sigma (St. Louis, MO, USA) |
| 2 | L-Homoserine lactone | Water | Santa Cruz Biotechnology | |
| 3 | α-Methyl-γ-butyrolactone | Water | Sigma (St. Louis, MO, USA) | |
| 4 | Furanone((Z-)-4-Bromo-5- (bromomethy-lene)-2(5H)-furanone) | First dissolved in ethanol, then diluted with water | Sigma (St. Louis, MO, USA) | |
| 5 | Vanillin | Group (2) Aromatic ring structures | Water | Sigma (St. Louis, MO, USA) |
| 6 | trans-Cinnamaldehyde | First dissolved in ethanol, then diluted with water | Sigma (St. Louis, MO, USA) | |
| 7 | Caffeic acid | First dissolved with ethanol, then diluted with water | Sigma (St. Louis, MO, USA) | |
| 8 | trans-Anethole | Water | Sigma (St. Louis, MO, USA) | |
| 9 | Capsaicin | First dissolved with ethanol, then diluted with water | Merck KGaA (Darmstadt, Germany) | |
| 10 | CAPE (caffeic acid phenethyl ester) | First dissolved with ethanol, then diluted with water | Merck KGaA (Darmstadt, Germany) | |
| 11 | Caffeine | Group (3) Heterocyclic compounds | water | Merck KGaA (Darmstadt, Germany) |
| 12 | Genipin | Water | Challenge Bioproducts Co., Ltd. | |
| 13 | Gardenoside | water | Nanjing Zelang Medical Technology Co.,Ltd | |
| 14 | PQS (2-heptyl-3-hydroxy-4-quinolone) | Group (4) Quinolone- and phenazine-based compounds relevant to QS systems of Pseudomonas spp | First dissolved with methanol, then diluted with water | Merck KGaA (Darmstadt, Germany) |
| 15 | MOQ (1H-2-methyl-4-quinolone) | First dissolved with methanol, then diluted with water | Prof. Fetzner’s 2 | |
| 16 | HOQ (1H-3-hydroxyl-4-quinolone) | First dissolved with methanol, then diluted with water | Prof. Fetzner’s 3 | |
| 17 | MHOQ (1H-2-methyl-3-hydroxyl-4-quinolone) | First dissolved with methanol, then diluted with water | Prof. Fetzner’s 4 | |
| 18 | PYO (pyocyanine) | First dissolved with methanol, then diluted with water | Merck KGaA (Darmstadt, Germany) | |
| 19 | PCA (Phenazine carboxylic acid) | First dissolved with methanol, then diluted with water | Key Organics Ltd. (Camelford, UK) | |
| 20 | PMS (Phenazine methosulfate) | First dissolved with methanol, then diluted with water | Sigma (St. Louis, MO, USA) | |
| 21 | Itaconic acid | Group (5) Structurally unrelated compounds | First dissolved in ethanol, then diluted with water | Sigma (St. Louis, MO, USA) |
| 22 | Polygodial | Water | Santa Cruz Biotechnology | |
| 23 | Berberine | First dissolved in ethanol, then diluted with water | Sigma (St. Louis, MO, USA) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, X.; Vila-Sanjurjo, C.; Singh, R.; Philipp, B.; Goycoolea, F.M. Screening of Bacterial Quorum Sensing Inhibitors in a Vibrio fischeri LuxR-Based Synthetic Fluorescent E. coli Biosensor. Pharmaceuticals 2020, 13, 263. https://doi.org/10.3390/ph13090263
Qin X, Vila-Sanjurjo C, Singh R, Philipp B, Goycoolea FM. Screening of Bacterial Quorum Sensing Inhibitors in a Vibrio fischeri LuxR-Based Synthetic Fluorescent E. coli Biosensor. Pharmaceuticals. 2020; 13(9):263. https://doi.org/10.3390/ph13090263
Chicago/Turabian StyleQin, Xiaofei, Celina Vila-Sanjurjo, Ratna Singh, Bodo Philipp, and Francisco M. Goycoolea. 2020. "Screening of Bacterial Quorum Sensing Inhibitors in a Vibrio fischeri LuxR-Based Synthetic Fluorescent E. coli Biosensor" Pharmaceuticals 13, no. 9: 263. https://doi.org/10.3390/ph13090263
APA StyleQin, X., Vila-Sanjurjo, C., Singh, R., Philipp, B., & Goycoolea, F. M. (2020). Screening of Bacterial Quorum Sensing Inhibitors in a Vibrio fischeri LuxR-Based Synthetic Fluorescent E. coli Biosensor. Pharmaceuticals, 13(9), 263. https://doi.org/10.3390/ph13090263