Adipose MSCs Suppress MCF7 and MDA-MB-231 Breast Cancer Metastasis and EMT Pathways Leading to Dormancy via Exosomal-miRNAs Following Co-Culture Interaction
Abstract
:1. Introduction
2. Results
2.1. Adipose MSC of Co-Culture Promotes Cancer Dormancy and Chemoresistance
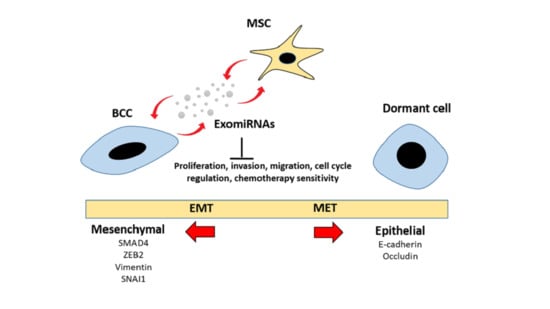
2.2. Adipose MSC of Co-Culture Suppresses Breast Cancer Metastasis through MET Transformation
2.3. Intracellular Transfer of Exosomes and RNAs from Adipose MSC to Breast Cancer Cells Facilitate Dormancy Acquisition and Metastasis Inhibition
2.4. The Distribution of Small RNAs Differs between Cells and Exosomes
2.5. Alteration of miRNAs Expression Profile in Cells and Exosomes Following Co-Culture
| Up/Down-Regulated miRNA | Exo-MSC Fold Change >2 | p-Value | Cell-MCF7 Fold Change >2 | p-Value |
|---|---|---|---|---|
| hsa-miR-200a-5p 1 | 4.99 | 0.006 | 9.84 | 0.000 |
| hsa-miR-203a-3p | 3.01 | 0.179 | 8.24 | 0.000 |
| hsa-miR-941 1 | 3.10 | 0.066 | 7.62 | 0.005 |
| hsa-miR-200a-3p | 3.50 | 0.062 | 6.93 | 0.000 |
| hsa-miR-629-5p 1 | 2.89 | 0.100 | 6.86 | 0.000 |
| hsa-miR-200b-3p | 4.43 | 0.002 | 6.81 | 0.000 |
| hsa-miR-589-5p | 2.00 | 0.441 | 6.17 | 0.002 |
| hsa-miR-200c-3p | 5.30 | 0.012 | 5.69 | 0.002 |
| hsa-miR-3615 | 3.32 | 0.122 | 5.60 | 0.002 |
| hsa-miR-7-5p | 3.20 | 0.005 | 4.73 | 0.000 |
| hsa-miR-185-5p | 3.91 | 0.017 | 3.65 | 0.001 |
| hsa-miR-1268b | 2.22 | 0.315 | 3.57 | 0.058 |
| hsa-miR-1268a | 2.22 | 0.315 | 3.57 | 0.058 |
| hsa-miR-375 | 3.76 | 0.045 | 3.41 | 0.018 |
| hsa-miR-222-5p | −2.06 | 0.232 | −5.18 | 0.001 |
| hsa-miR-10a-5p | −2.30 | 0.028 | −5.24 | 0.000 |
| hsa-miR-221-5p | −3.61 | 0.026 | −5.54 | 0.000 |
| hsa-miR-143-3p | −2.58 | 0.032 | −5.87 | 0.000 |
| hsa-miR-199b-3p | −2.00 | 0.259 | −6.19 | 0.000 |
| hsa-miR-199a-3p | −2.00 | 0.259 | −6.20 | 0.000 |
| hsa-miR-199a-5p | −2.31 | 0.173 | −7.06 | 0.000 |
| hsa-miR-100-5p | −2.28 | 0.000 | −7.19 | 0.000 |
| hsa-miR-486-5p 1 | −2.89 | 0.004 | −7.46 | 0.000 |
| hsa-miR-125b-1-3p | −2.00 | 0.000 | −7.89 | 0.000 |
| hsa-miR-155-5p | −13.68 | 0.000 | −9.34 | 0.000 |
| hsa-miR-10b-5p 1 | −2.80 | 0.011 | −10.22 | 0.000 |
| hsa-miR-224-5p | −3.30 | 0.001 | −23.77 | 0.000 |
| Up/down-regulated miRNA | Exo-MSC Fold change >2 | p-value | Cell-MDA-MB-231 Fold change >2 | p-value |
| hsa-miR-941 1 | 3.58 | 0.114 | 12.89 | 0.000 |
| hsa-miR-629-5p 1 | 4.82 | 0.006 | 9.61 | 0.000 |
| hsa-miR-146a-5p | 6.31 | 0.000 | 8.46 | 0.000 |
| hsa-miR-1180-3p | 4.13 | 0.128 | 8.39 | 0.001 |
| hsa-miR-1246 | 4.15 | 0.008 | 7.64 | 0.000 |
| hsa-miR-1290 | 4.74 | 0.027 | 7.46 | 0.002 |
| hsa-miR-200a-5p 1 | 3.05 | 0.150 | 6.85 | 0.000 |
| hsa-miR-1301-3p | 3.07 | 0.308 | 6.04 | 0.034 |
| hsa-miR-7704 | 6.22 | 0.000 | 6 | 0.002 |
| hsa-miR-486-5p 1 | −2.72 | 0.052 | −6.97 | 0.000 |
| hsa-miR-10b-5p 1 | −3.93 | 0.004 | −11.16 | 0.000 |
| Target Gene in MCF7 | Gene Description | Dysregulated miRNAs | O/E Ratio |
|---|---|---|---|
| ACVR2A | Activin A receptor, type IIA | 10 | 0.916248 |
| CBL | Cbl proto-oncogene, E3 ubiquitin protein ligase | 8 | 0.52331 |
| CAPRIN1 1 | Cell cycle-associated protein 1 | 7 | 0.562918 |
| MAP3K7 1 | Mitogen-activated protein kinase kinase kinase 7 | 6 | 1.13509 |
| CDC42 | Cell division cycle 42 | 6 | 0.777232 |
| PTEN 1 | Phosphatase and tensin homolog | 6 | 0.633646 |
| SMAD2 1 | SMAD family member 2 | 6 | 0.400451 |
| IGF1R | Insulin-like growth factor 1 receptor | 6 | 0.392482 |
| ERBB4 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 | 6 | 0.372997 |
| NFAT5 1 | Nuclear factor of activated T-cells 5, tonicity-responsive | 6 | 0.308160 |
| Target gene in MDA-MB-231 | Gene description | Dysregulated miRNAs | O/E ratio |
| CADM1 | cell adhesion molecule 1 | 3 | 0.841896 |
| NFAT5 1 | nuclear factor of activated T-cells 5, tonicity-responsive | 3 | 0.378196 |
| GAB1 | GRB2-associated binding protein 1 | 2 | 0.977964 |
| MAP3K7 1 | mitogen-activated protein kinase kinase kinase 7 | 2 | 0.92871 |
| BRCA1 | breast cancer 1, early onset | 2 | 0.768397 |
| PTEN 1 | phosphatase and tensin homolog | 2 | 0.518440 |
| ARHGAP5 | Rho GTPase activating protein 5 | 2 | 0.494601 |
| CD28 | CD28 molecule | 2 | 0.428873 |
| CAPRIN1 1 | cell cycle associated protein 1 | 2 | 0.394774 |
| SMAD2 1 | SMAD family member 2 | 2 | 0.327642 |

2.6. Biological Pathway Potentially Influenced by Dysregulated miRNAs-Mediated Breast Cancer Cells Dormancy
2.7. miR-941 Is Specifically Upregulated in Co-Culture Exosomes and BCCs
2.8. miR-941 Overexpression Suppress BCCs Viability and Metastasis via MET Regulations
3. Discussion
4. Materials and Methods
4.1. Human Breast Cancer Cells Lines
4.2. Adipose Mesenchymal Stem Cells
4.3. Transwell Indirect Co-Culture Assay
4.4. Internalization of Exosome and RNAs
4.5. Cell Proliferation Assay
4.6. Cell Cycle Analysis
4.7. Transwell Invasion and Migration Assays
4.8. Wound Healing Assay
4.9. Chemoresistant MTT Assay
4.10. 3D Spheres Formation Assay
4.11. Flow Cytometric Analysis of CD24 and CD44 Surface Markers
4.12. Isolation of Exosomes
4.13. Exosomes Characterization
4.14. Exosomes Functionality Study
4.15. MicroRNA Expression Study
4.15.1. Isolation of Cellular and Exosomal Total RNAs
4.15.2. Construction of Small RNA Libraries and Sequencing
4.15.3. Analysis of miRNA Sequencing Data
4.15.4. Prediction of Highly Dysregulated miRNAs Target Genes and Pathway Enrichment Analysis
4.15.5. Verification of miRNA NGS Results by qRT-PCR
4.15.6. Changes in the Expression of MDR, CSC, DNA Repair, Epithelial and Mesenchymal Genes
4.15.7. Transfection of miRNA Mimic
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Sakha, S.; Muramatsu, T.; Ueda, K.; Inazawa, J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci. Rep. 2016, 6, 38750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redig, A.J.; McAllister, S.S. Breast cancer as a systemic disease: A view of metastasis. J. Intern. Med. 2013, 274, 113–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Ye, P.; Long, X. Differential Expression Profiles of the Transcriptome in Breast Cancer Cell Lines Revealed by Next Generation Sequencing. Cell. Physiol. Biochem. 2017, 44, 804–816. [Google Scholar] [CrossRef]
- Plava, J.; Cihova, M.; Burikova, M.; Matuskova, M.; Kucerova, L.; Miklikova, S. Recent advances in understanding tumor stroma-mediated chemoresistance in breast cancer. Mol. Cancer 2019, 18, 1–10. [Google Scholar] [CrossRef]
- Tang, Y.-T.; Huang, Y.-Y.; Li, J.-H.; Qin, S.-H.; Xu, Y.; An, T.-X.; Liu, C.-C.; Wang, Q.; Zheng, L. Alterations in exosomal miRNA profile upon epithelial-mesenchymal transition in human lung cancer cell lines. BMC Genom. 2018, 19, 802. [Google Scholar] [CrossRef]
- Hu, W.; Tan, C.; He, Y.; Zhang, G.; Xu, Y.; Tang, J. Functional miRNAs in breast cancer drug resistance. Onco Targets Ther. 2018, 11, 1529–1541. [Google Scholar] [CrossRef] [Green Version]
- Shao, S.; Zhao, X.; Zhang, X.; Luo, M.; Zuo, X.; Huang, S.; Wang, Y.; Gu, S.; Zhao, X. Notch1 signaling regulates the epithelial–mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol. Cancer 2015, 14, 28. [Google Scholar] [CrossRef] [Green Version]
- Yadav, A.S.; Pandey, P.R.; Butti, R.; Radharani, N.N.V.; Roy, S.; Bhalara, S.R.; Gorain, M.; Kundu, G.C.; Kumar, D. The Biology and Therapeutic Implications of Tumor Dormancy and Reactivation. Front. Oncol. 2018, 8, 72. [Google Scholar] [CrossRef] [Green Version]
- Ramdasi, S.; Sarang, S.; Viswanathan, C. Potential of Mesenchymal Stem Cell based application in Cancer. Int. J. Hematol. Stem Cell Res. 2015, 9, 95–103. [Google Scholar]
- Reza, A.M.M.T.; Choi, Y.-J.; Yasuda, H.; Kim, J.-H. Human adipose mesenchymal stem cell-derived exosomal-mirnas are critical factors for inducing anti-proliferation signalling to a2780 and skov-3 ovarian cancer cells. Sci. Reports 2016, 6, 38498. [Google Scholar] [CrossRef]
- Ono, M.; Kosaka, N.; Tominaga, N.; Yoshioka, Y.; Takeshita, F.; Takahashi, R.-U.; Yoshida, M.; Tsuda, H.; Tamura, K.; Ochiya, T. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 2014, 7, ra63. [Google Scholar] [CrossRef]
- Kucerova, L.; Skolekova, S.; Matuskova, M.; Bohac, M.; Kozovska, Z. Altered features and increased chemosensitivity of human breast cancer cells mediated by adipose tissue-derived mesenchymal stromal cells. BMC Cancer 2013, 13, 535. [Google Scholar] [CrossRef] [Green Version]
- Cocce, V.; Balducci, L.; Falchetti, M.L.; Pascucci, L.; Ciusani, E.; Brini, A.T.; Sisto, F.; Piovani, G.; Alessandri, G.; Parati, E.; et al. Fluorescent immortalized human adipose derived stromal cells (hascs-ts/gfp+) for studying cell drug delivery mediated by microvesicles. Anticancer Agents Med. Chem. 2017, 17, 1578–1585. [Google Scholar] [CrossRef]
- Gelao, L.; Criscitiello, C.; Fumagalli, L.; Locatelli, M.; Manunta, S.; Esposito, A.; Minchella, I.; Goldhirsch, A.; Curigliano, G. Tumour dormancy and clinical implications in breast cancer. Ecancermedicalscience 2013, 7, 320. [Google Scholar]
- Zhang, P.-P.; Wang, X.-L.; Zhao, W.; Qi, B.; Yang, Q.; Wan, H.-Y.; Shuang, Z.-Y.; Liu, M.; Li, X.; Li, S.-P.; et al. DNA Methylation-mediated Repression of miR-941 Enhances Lysine (K)-specific Demethylase 6B Expression in Hepatoma Cells. J. Biol. Chem. 2014, 289, 24724–24735. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-G.; Kim, T.-O.; Bae, J.-H.; Shim, J.-W.; Kang, M.J.; Yang, K.; Ting, A.H.; Yi, J.M. Epigenetically regulatedMIR941andMIR1247target gastric cancer cell growth and migration. Epigenetics 2014, 9, 1018–1030. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; He, Y.; Wang, X.; Lu, F.; Gao, J. Adipose-derived mesenchymal stem cells exhibit tumor tropism and promote tumorsphere formation of breast cancer cells. Oncol. Rep. 2019, 41, 2126–2136. [Google Scholar] [CrossRef] [Green Version]
- Koh, B.; Jeon, H.; Kim, D.; Kang, D.; Kim, K.R. Effect of fibroblast co-culture on the proliferation, viability and drug response of colon cancer cells. Oncol. Lett. 2018, 17, 2409–2417. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.J.; Oh, M.-S.; Lee, D.W.; Kuh, H.-J. Multiplex quantitative analysis of stroma-mediated cancer cell invasion, matrix remodeling, and drug response in a 3D co-culture model of pancreatic tumor spheroids and stellate cells. J. Exp. Clin. Cancer Res. 2019, 38, 258. [Google Scholar] [CrossRef] [Green Version]
- Bartosh, T.J.; Ullah, M.; Zeitouni, S.; Beaver, J.; Prockop, D.J. Cancer cells enter dormancy after cannibalizing mesenchymal stem/stromal cells (MSCs). Proc. Natl. Acad. Sci. USA 2016, 113, E6447–E6456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bliss, S.A.; Sinha, G.; Sandiford, O.A.; Williams, L.M.; Engelberth, D.J.; Guiro, K.; Isenalumhe, L.L.; Greco, S.J.; Ayer, S.; Bryan, M.; et al. Mesenchymal Stem Cell–Derived Exosomes Stimulate Cycling Quiescence and Early Breast Cancer Dormancy in Bone Marrow. Cancer Res. 2016, 76, 5832–5844. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Su, X.; Xu, M.; Xiao, X.; Li, X.; Li, H.; Keating, A.; Zhao, R.C. Exosomes secreted by mesenchymal stromal/stem cell-derived adipocytes promote breast cancer cell growth via activation of Hippo signaling pathway. Stem Cell Res. Ther. 2019, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Farahmand, L.; Esmaeili, R.; Eini, L.; Majidzadeh-A, K. The effect of mesenchymal stem cell-conditioned medium on proliferation and apoptosis of breast cancer cell line. J. Cancer Res. Ther. 2018, 14, 341–344. [Google Scholar]
- Marlow, R.; Honeth, G.; Lombardi, S.; Cariati, M.; Hessey, S.; Pipili, A.; Mariotti, V.; Buchupalli, B.; Foster, K.; Bonnet, D.; et al. A Novel Model of Dormancy for Bone Metastatic Breast Cancer Cells. Cancer Res. 2013, 73, 6886–6899. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Zhou, C.; Chen, X.; Tao, C.; Cheng, H.; Lu, X. Suppression of tumor cell proliferation and migration by human umbilical cord mesenchymal stem cells: A possible role for apoptosis and Wnt signaling. Oncol. Lett. 2018, 15, 8536–8544. [Google Scholar] [CrossRef]
- Boo, L.; Ho, W.Y.; Ali, N.M.; Yeap, S.K.; Ky, H.; Chan, K.G.; Yin, W.F.; Satharasinghe, D.A.; Liew, W.C.; Tan, S.W.; et al. Phenotypic and microRNA transcriptomic profiling of the MDA-MB-231 spheroid-enriched CSCs with comparison of MCF-7 microRNA profiling dataset. PeerJ 2017, 5, 3551. [Google Scholar] [CrossRef] [Green Version]
- Ignatov, A.; Eggemann, H.; Burger, E.; Ignatov, T. Patterns of breast cancer relapse in accordance to biological subtype. J. Cancer Res. Clin. Oncol. 2018, 144, 1347–1355. [Google Scholar] [CrossRef]
- Dioufa, N.; Clark, A.M.; Ma, B.; Beckwitt, C.H.; Wells, A. Bi-directional exosome-driven intercommunication between the hepatic niche and cancer cells. Mol. Cancer 2017, 16, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Scioli, M.G.; Storti, G.; D’Amico, F.; Gentile, P.; Kim, B.-S.; Cervelli, V.; Orlandi, A. Adipose-Derived Stem Cells in Cancer Progression: New Perspectives and Opportunities. Int. J. Mol. Sci. 2019, 20, 3296. [Google Scholar] [CrossRef] [Green Version]
- Roche, J. The Epithelial-to-Mesenchymal Transition in Cancer. Cancers 2018, 10, 52. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhang, L. Members of the microRNA-200 family are promising therapeutic targets in cancer. Exp. Ther. Med. 2017, 14, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.-D.; Lv, M.; Chen, W.-X.; Zhong, S.-L.; Zhang, X.-H.; Chen, L.; Ma, T.-F.; Tang, J.-H.; Zhao, J.-H. Role of miR-155 in drug resistance of breast cancer. Tumor Biol. 2015, 36, 1395–1401. [Google Scholar] [CrossRef]
- Yin, C.; Zhang, G.; Sun, R.; Pan, X.; Wang, X.; Li, H.; Sun, Y. miR-185-5p inhibits F-actin polymerization and reverses epithelial mesenchymal transition of human breast cancer cells by modulating RAGE. Mol. Med. Rep. 2018, 18, 2621–2630. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.-X.; Bradbury, R.; Flamini, V.; Wu, B.; Jordan, N.; Jiang, W.G. MicroRNA-7 suppresses the homing and migration potential of human endothelial cells to highly metastatic human breast cancer cells. Br. J. Cancer 2017, 117, 89–101. [Google Scholar] [CrossRef] [Green Version]
- Si, C.; Yu, Q.; Yao, Y. Effect of miR-146a-5p on proliferation and metastasis of triple-negative breast cancer via regulation of SOX5. Exp. Ther. Med. 2018, 15, 4515–4521. [Google Scholar] [CrossRef] [Green Version]
- Iacona, J.R.; Lutz, C.S. Mir-146a-5p: Expression, regulation, and functions in cancer. Wiley interdisciplinary reviews. RNA 2019, 10, e1533. [Google Scholar]
- Yan, G.; Li, C.; Zhao, Y.; Yue, M.; Wang, L. Downregulation of microRNA-629-5p in colorectal cancer and prevention of the malignant phenotype by direct targeting of low-density lipoprotein receptor-related protein 6. Int. J. Mol. Med. 2019, 44, 1139–1150. [Google Scholar] [CrossRef]
- Loh, H.-Y.; Norman, B.P.; Lai, K.-S.; Rahman, N.M.A.N.A.; Alitheen, N.B.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef] [Green Version]
- Elkhouly, A.M.; Youness, R.A.; Gad, M.Z. MicroRNA-486-5p and microRNA-486-3p: Multifaceted pleiotropic mediators in oncological and non-oncological conditions. Non-Coding RNA Res. 2020, 5, 11–21. [Google Scholar] [CrossRef]
- Sheedy, P.; Medarova, Z. The fundamental role of miR-10b in metastatic cancer. Am. J. Cancer Res. 2018, 8, 1674–1688. [Google Scholar]
- Yu, S.-J.; Yang, L.; Gen-Hong, D.; Kuang, X.-Y.; Di, G.-H.; Shao, Z.-M. MicroRNA-200a confers chemoresistance by antagonizing TP53INP1 and YAP1 in human breast cancer. BMC Cancer 2018, 18, 1–11. [Google Scholar] [CrossRef]
- Prunier, C.; Baker, D.; Ten-Dijke, P.; Ritsma, L. Tgf-beta family signaling pathways in cellular dormancy. Trends Cancer 2019, 5, 66–78. [Google Scholar] [CrossRef]
- Li, N.Y.; Weber, C.E.; Wai, P.Y.; Cuevas, B.D.; Zhang, J.; Kuo, P.C.; Mi, Z. An MAPK-dependent pathway induces epithelial-mesenchymal transition via Twist activation in human breast cancer cell lines. Surgery 2013, 154, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Triaca, V.; Carito, V.; Fico, E.; Rosso, P.; Fiore, M.; Ralli, M.; Lambiase, A.; Greco, A.; Tirassa, P. Cancer stem cells-driven tumor growth and immune escape: The Janus face of neurotrophins. Aging 2019, 11, 11770–11792. [Google Scholar] [CrossRef]
- Appert-Collin, A.; Hubert, P.; Crémel, G.; Bennasroune, A. Role of ErbB Receptors in Cancer Cell Migration and Invasion. Front. Pharmacol. 2015, 6, 283. [Google Scholar] [CrossRef] [Green Version]
- Hopkins, B.D.; Goncalves, M.D.; Cantley, L.C. Insulin—PI3K signalling: An evolutionarily insulated metabolic driver of cancer. Nat. Rev. Endocrinol. 2020, 16, 276–283. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.; Zhang, W.; Zheng, Y.; Wang, Z.; Liu, L.; Wu, H.; Ye, J.; Qi, B.; Wu, Y.; et al. Axon guidance molecule semaphorin3A is a novel tumor suppressor in head and neck squamous cell carcinoma. Oncotarget 2016, 7, 6048–6062. [Google Scholar] [CrossRef]
- Luo, M.; Guan, J.L. Focal adhesion kinase: A prominent determinant in breast cancer initiation, progression and metastasis. Cancer Lett. 2010, 289, 127–139. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Zhao, J.; Li, H.; He, K.; Chen, S.-H.; Mayer, L.; Unkeless, J.C.; Xiong, H. Toll-Like Receptors on Tumor Cells Facilitate Evasion of Immune Surveillance. Cancer Res. 2005, 65, 5009–5014. [Google Scholar] [CrossRef] [Green Version]
- Romero, I.; Garrido, F.; Garcia-Lora, A.M. Metastases in Immune-Mediated Dormancy: A New Opportunity for Targeting Cancer. Cancer Res. 2014, 74, 6750–6757. [Google Scholar] [CrossRef] [Green Version]
- van Staveren, W.; Solís, D.W.; Hebrant, A.; Detours, V.; Dumont, J.E.; Maenhaut, C. Human cancer cell lines: Experimental models for cancer cells in situ? For cancer stem cells? Biochim. Biophys. Acta Rev. Cancer 2009, 1795, 92–103. [Google Scholar] [CrossRef]
- Verma, M. Personalized Medicine and Cancer. J. Pers. Med. 2012, 2, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Schweizer, R.; Tsuji, W.; Gorantla, V.S.; Marra, K.G.; Rubin, J.P.; Plock, J.A. The Role of Adipose-Derived Stem Cells in Breast Cancer Progression and Metastasis. Stem Cells Int. 2015, 2015, 1–17. [Google Scholar] [CrossRef]
- Ali, N.M.; Boo, L.; Yeap, S.K.; Ky, H.; Satharasinghe, D.A.; Liew, W.C.; Ong, H.K.; Cheong, S.K.; Kamarul, T. Probable impact of age and hypoxia on proliferation and microRNA expression profile of bone marrow-derived human mesenchymal stem cells. PeerJ 2016, 4, e1536. [Google Scholar] [CrossRef]
- Santos, J.C.; Lima, N.D.S.; Sarian, L.O.; Matheu, A.; Ribeiro, M.L.; Derchain, S.F.M. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Gordon, A.; Hannon, G. Fastx-Toolkit. FASTQ/A Short Reads Preprocessing Tools (unpublished). Available online: http://hannonlab.cshl.edu/fastx_toolkit (accessed on 25 May 2018).
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.-P.; Lee, C.-Y.; Tsai, M.-H.; Chiu, Y.-C.; Hsiao, C.K.; Lai, L.-C.; Chuang, E.Y. miRSystem: An Integrated System for Characterizing Enriched Functions and Pathways of MicroRNA Targets. PLoS ONE 2012, 7, e42390. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]







| miRNA | Sequence Accession Number | Target Sequence |
|---|---|---|
| hsa-miR-146a-5p | MIMAT0000449 | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-941 | MIMAT0004984 | CACCCGGCUGUGUGCACAUGUGC |
| hsa-miR-10b-5p | MIMAT0000254 | UACCCUGUAGAACCGAAUUUGUG |
| hsa-miR-760 | MIMAT0004957 | CGGCUCUGGGUCUGUGGGGA |
| hsa-miR-205-5p | MIMAT0000266 | UCCUUCAUUCCACCGGAGUCUG |
| Genes | Target Sequence (5′-3′)/ Assay ID |
|---|---|
| Housekeeping Reference Genes | |
| ACTB | FP: 5′-AGAGCTACGAGCTGCCTGAC-3′ RP: 5′-AGCACTGTGTTGGCGTACAG-3′ |
| Epithelial genes | |
| E-cadherin | FP: 5′-ACAGGAACACAGGAGTCATCAG-3′ RP: 5′- CCCTTGTACGTGGTGGGATT-3′ |
| OCLN | FP: 5′-TGCCTAGCTACCCCCATCTT-3′ RP: 5′-TGC ACC CAG CACAGATCAAT-3′ |
| Mesenchymal Genes | |
| SNAIL | FP: 5′-AGTGGTTCTTCTGCGCTACTG-3′ RP: 5′-TGCTGGAAGGTAAACTCTGGATTAG-3′ |
| SMAD4 | FP: 5′-CTCCAGCTATCAGTCTGTCA-3′ RP: 5′-GATGCTCTGTCTTGGGTAATC-3′ |
| Vimentin | FP: 5′-CCTGCAATCTTTCAGACAGG-3′ RP: 5′-CTCCTGGATTTCCTCTTCGT-3′ |
| Zeb2 | FP: 5′-TTTCAGGGAGAATTGCTTGA-3′ RP: 5′-CACATGCATACATGCCACTC-3′ |
| Multidrug-resistant-ABC Transporter Genes | |
| ABCC2 | FP: 5′-TGC AGC CTC CAT AAC CAT GAG-3′ RP: 5′-GAT GCC TGC CAT TGG ACC TA-3′ |
| ABCG2 | FP: 5′-CAG GTC TGT TGG TCA ATC TCA CA-3′ RP: 5′-TCC ATA TCG TGG AAT GCT GAA G-3′ |
| ABCA3 | FP: 5′-CAA AAC CCT GGA TCA CGT GTT-3′ RP: 5′-CCT CCG CGT CTC GTA GTT CT-3′ |
| Cancer Stem Cell Genes | |
| ALDH1A1 | FP: 5′-AGCAGGAGTGTTTACCAAAGA-3′ RP: 5′-CCCAGTTCTCTTCCATTTCCAG-3′ |
| CD44 | FP: 5′-CACAAATGGCTGGTACGTCTT-3′ RP: 5′-TTCATCTTCATTTTCTTCATTTGG-3′ |
| DNA Repair Genes (TaqMan® Probe) | |
| PARP1 | Hs00242302_m1 |
| PARP3 | Hs00193946_m1 |
| MLH1 | Hs00979919_m1 |
| CCND2 | Hs00153380_m1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohd Ali, N.; Yeap, S.K.; Ho, W.Y.; Boo, L.; Ky, H.; Satharasinghe, D.A.; Tan, S.W.; Cheong, S.K.; Huang, H.D.; Lan, K.C.; et al. Adipose MSCs Suppress MCF7 and MDA-MB-231 Breast Cancer Metastasis and EMT Pathways Leading to Dormancy via Exosomal-miRNAs Following Co-Culture Interaction. Pharmaceuticals 2021, 14, 8. https://doi.org/10.3390/ph14010008
Mohd Ali N, Yeap SK, Ho WY, Boo L, Ky H, Satharasinghe DA, Tan SW, Cheong SK, Huang HD, Lan KC, et al. Adipose MSCs Suppress MCF7 and MDA-MB-231 Breast Cancer Metastasis and EMT Pathways Leading to Dormancy via Exosomal-miRNAs Following Co-Culture Interaction. Pharmaceuticals. 2021; 14(1):8. https://doi.org/10.3390/ph14010008
Chicago/Turabian StyleMohd Ali, Norlaily, Swee Keong Yeap, Wan Yong Ho, Lily Boo, Huynh Ky, Dilan Amila Satharasinghe, Sheau Wei Tan, Soon Keng Cheong, Hsien Da Huang, Kuan Chun Lan, and et al. 2021. "Adipose MSCs Suppress MCF7 and MDA-MB-231 Breast Cancer Metastasis and EMT Pathways Leading to Dormancy via Exosomal-miRNAs Following Co-Culture Interaction" Pharmaceuticals 14, no. 1: 8. https://doi.org/10.3390/ph14010008
APA StyleMohd Ali, N., Yeap, S. K., Ho, W. Y., Boo, L., Ky, H., Satharasinghe, D. A., Tan, S. W., Cheong, S. K., Huang, H. D., Lan, K. C., Chiew, M. Y., & Ong, H. K. (2021). Adipose MSCs Suppress MCF7 and MDA-MB-231 Breast Cancer Metastasis and EMT Pathways Leading to Dormancy via Exosomal-miRNAs Following Co-Culture Interaction. Pharmaceuticals, 14(1), 8. https://doi.org/10.3390/ph14010008