DNA Aptamers against Vaccinia-Related Kinase (VRK) 1 Block Proliferation in MCF7 Breast Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. Selection of High-Affinity Aptamers against VRK1
2.2. Bioinformatic Analysis of the Secondary and Tertiary Structures
2.3. Characterization of the Aptamers Obtained against VRK1
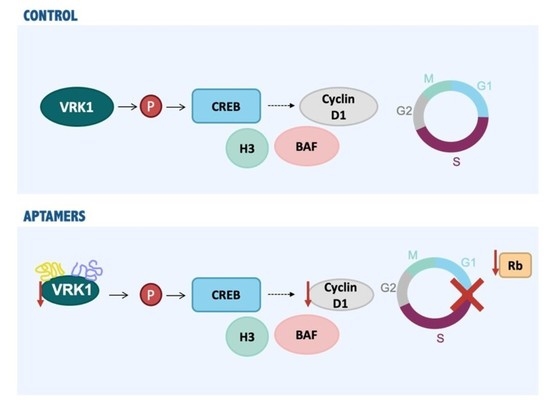
2.4. VRK1 Aptamers Block Proliferation in MCF7 Breast Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. In Vitro Selection Procedure
4.3. Enzyme-Linked Oligonucleotide Assay
4.4. Aptamer Cloning, Sequencing and Secondary and Tertiary Structure Prediction
4.5. Aptamer Stability Assay
4.6. Slotblot
4.7. Aptamer Quantification
4.8. Cell Culture and Transfection
4.9. Protein Extraction, Dodecyl Sulphate-Polyacrilamide Gel Electrophoresis and Immunoblotting
4.10. MTT Assay
4.11. Cell Cycle Assay
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [Green Version]
- Nezu, J.; Oku, A.; Jones, M.H.; Shimane, M. Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with structural similarity to vaccinia virus B1R kinase. Genomics 1997, 45, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Nichols, R.J.; Traktman, P. Characterization of three paralogous members of the Mammalian vaccinia related kinase family. J. Biol. Chem. 2004, 279, 7934–7946. [Google Scholar] [CrossRef] [Green Version]
- Kang, T.H.; Park, D.Y.; Choi, Y.H.; Kim, K.J.; Yoon, H.S.; Kim, K.T. Mitotic histone H3 phosphorylation by vaccinia-related kinase 1 in mammalian cells. Mol. Cell. Biol. 2007, 27, 8533–8546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz-Garcia, M.; Monsalve, D.M.; Sevilla, A.; Lazo, P.A. Vaccinia-related kinase 1 (VRK1) is an upstream nucleosomal kinase required for the assembly of 53BP1 foci in response to ionizing radiation-induced DNA damage. J. Biol. Chem. 2012, 287, 23757–23768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monsalve, D.M.; Campillo-Marcos, I.; Salzano, M.; Sanz-Garcia, M.; Cantarero, L.; Lazo, P.A. VRK1 phosphorylates and protects NBS1 from ubiquitination and proteasomal degradation in response to DNA damage. Biochim. Biophys. Acta 2016, 1863, 760–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salzano, M.; Sanz-Garcia, M.; Monsalve, D.M.; Moura, D.S.; Lazo, P.A. VRK1 chromatin kinase phosphorylates H2AX and is required for foci formation induced by DNA damage. Epigenetics 2015, 10, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, A.J.; Rao, V.A.; Pommier, Y.; Bonner, W.M. The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks. Cell Cycle 2010, 9, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sanchez, I.; Valbuena, A.; Vazquez-Cedeira, M.; Khadake, J.; Sanz-Garcia, M.; Carrillo-Jimenez, A.; Lazo, P.A. VRK1 interacts with p53 forming a basal complex that is activated by UV-induced DNA damage. FEBS Lett. 2014, 588, 692–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vega, F.M.; Sevilla, A.; Lazo, P.A. p53 Stabilization and accumulation induced by human vaccinia-related kinase 1. Mol. Cell Biol. 2004, 24, 10366–10380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campillo-Marcos, I.; Lazo, P.A. Implication of the VRK1 chromatin kinase in the signaling responses to DNA damage: A therapeutic target? Cell. Mol. Life Sci. 2018, 75, 2375–2388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valbuena, A.; Sanz-Garcia, M.; Lopez-Sanchez, I.; Vega, F.M.; Lazo, P.A. Roles of VRK1 as a new player in the control of biological processes required for cell division. Cell Signal. 2011, 23, 1267–1272. [Google Scholar] [CrossRef] [Green Version]
- Kang, T.H.; Park, D.Y.; Kim, W.; Kim, K.T. VRK1 phosphorylates CREB and mediates CCND1 expression. J. Cell Sci. 2008, 121, 3035–3041. [Google Scholar] [CrossRef] [Green Version]
- Molitor, T.P.; Traktman, P. Depletion of the protein kinase VRK1 disrupts nuclear envelope morphology and leads to BAF retention on mitotic chromosomes. Mol. Biol. Cell 2014, 25, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Nichols, R.J.; Wiebe, M.S.; Traktman, P. The vaccinia-related kinases phosphorylate the N’ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol. Biol. Cell 2006, 17, 2451–2464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashiguchi, T.; Arakawa, S.; Takahashi, S.; Gonzalez, F.J.; Sueyoshi, T.; Negishi, M. Phosphorylation of Farnesoid X Receptor at Serine 154 Links Ligand Activation With Degradation. Mol. Endocrinol. 2016, 30, 1070–1080. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Sanchez, I.; Sanz-Garcia, M.; Lazo, P.A. Plk3 interacts with and specifically phosphorylates VRK1 in Ser342, a downstream target in a pathway that induces Golgi fragmentation. Mol. Cell. Biol. 2009, 29, 1189–1201. [Google Scholar] [CrossRef] [Green Version]
- Moura, D.S.; Fernandez, I.F.; Marin-Royo, G.; Lopez-Sanchez, I.; Martin-Doncel, E.; Vega, F.M.; Lazo, P.A. Oncogenic Sox2 regulates and cooperates with VRK1 in cell cycle progression and differentiation. Sci. Rep. 2016, 6, 28532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sevilla, A.; Santos, C.R.; Barcia, R.; Vega, F.M.; Lazo, P.A. c-Jun phosphorylation by the human vaccinia-related kinase 1 (VRK1) and its cooperation with the N-terminal kinase of c-Jun (JNK). Oncogene 2004, 23, 8950–8958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sevilla, A.; Santos, C.R.; Vega, F.M.; Lazo, P.A. Human vaccinia-related kinase 1 (VRK1) activates the ATF2 transcriptional activity by novel phosphorylation on Thr-73 and Ser-62 and cooperates with JNK. J. Biol. Chem. 2004, 279, 27458–27465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valbuena, A.; Lopez-Sanchez, I.; Lazo, P.A. Human VRK1 is an early response gene and its loss causes a block in cell cycle progression. PLoS ONE 2008, 3, e1642. [Google Scholar] [CrossRef] [PubMed]
- Wiebe, M.S.; Nichols, R.J.; Molitor, T.P.; Lindgren, J.K.; Traktman, P. Mice deficient in the serine/threonine protein kinase VRK1 are infertile due to a progressive loss of spermatogonia. Biol. Reprod. 2010, 82, 182–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molitor, T.P.; Traktman, P. Molecular genetic analysis of VRK1 in mammary epithelial cells: Depletion slows proliferation in vitro and tumor growth and metastasis in vivo. Oncogenesis 2013, 2, e48. [Google Scholar] [CrossRef] [Green Version]
- Santos, C.R.; Rodriguez-Pinilla, M.; Vega, F.M.; Rodriguez-Peralto, J.L.; Blanco, S.; Sevilla, A.; Valbuena, A.; Hernandez, T.; van Wijnen, A.J.; Li, F.; et al. VRK1 signaling pathway in the context of the proliferation phenotype in head and neck squamous cell carcinoma. Mol. Cancer Res. 2006, 4, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Valbuena, A.; Suarez-Gauthier, A.; Lopez-Rios, F.; Lopez-Encuentra, A.; Blanco, S.; Fernandez, P.L.; Sanchez-Cespedes, M.; Lazo, P.A. Alteration of the VRK1-p53 autoregulatory loop in human lung carcinomas. Lung Cancer 2007, 58, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Cui, X.; Chen, Y.; Shao, M.; Shao, X.; Shen, Y.; Liu, Q.; Wu, M.; Liu, J.; Ni, W.; et al. High VRK1 expression contributes to cell proliferation and survival in hepatocellular carcinoma. Pathol. Res. Pract. 2016, 212, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Kwon, J.H.; Kim, Y.B.; Kim, S.H.; Park, S.J.; Xu, W.; Jung, H.Y.; Kim, K.T.; Wang, H.J.; Choi, K.Y. Vaccinia-related kinase 1 promotes hepatocellular carcinoma by controlling the levels of cell cycle regulators associated with G1/S transition. Oncotarget 2015, 6, 30130–30148. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, T.; Pei, L.; Jing, J.; Hu, W.; Sun, T.; Liu, H. Expression of VRK1 and the downstream gene BANF1 in esophageal cancer. Biomed. Pharmacother. 2017, 89, 1086–1091. [Google Scholar] [CrossRef]
- Ben, Z.; Gong, L.; Qiu, Y. High expression of VRK1 is related to poor prognosis in glioma. Pathol. Res. Pract. 2018, 214, 112–118. [Google Scholar] [CrossRef]
- Fournier, M.V.; Martin, K.J.; Kenny, P.A.; Xhaja, K.; Bosch, I.; Yaswen, P.; Bissell, M.J. Gene expression signature in organized and growth-arrested mammary acini predicts good outcome in breast cancer. Cancer Res. 2006, 66, 7095–7102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mon, A.M.; MacKinnon, A.C., Jr.; Traktman, P. Overexpression of the VRK1 kinase, which is associated with breast cancer, induces a mesenchymal to epithelial transition in mammary epithelial cells. PLoS ONE 2018, 13, e0203397. [Google Scholar] [CrossRef]
- Finetti, P.; Cervera, N.; Charafe-Jauffret, E.; Chabannon, C.; Charpin, C.; Chaffanet, M.; Jacquemier, J.; Viens, P.; Birnbaum, D.; Bertucci, F. Sixteen-kinase gene expression identifies luminal breast cancers with poor prognosis. Cancer Res. 2008, 68, 767–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, K.J.; Patrick, D.R.; Bissell, M.J.; Fournier, M.V. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS ONE 2008, 3, e2994. [Google Scholar] [CrossRef] [Green Version]
- Salzano, M.; Vazquez-Cedeira, M.; Sanz-Garcia, M.; Valbuena, A.; Blanco, S.; Fernandez, I.F.; Lazo, P.A. Vaccinia-related kinase 1 (VRK1) confers resistance to DNA-damaging agents in human breast cancer by affecting DNA damage response. Oncotarget 2014, 5, 1770–1778. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 1992, 355, 850–852. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef]
- Thiel, K.W.; Giangrande, P.H. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides 2009, 19, 209–222. [Google Scholar] [CrossRef]
- Amani, N.; Shokrzadeh, M.; Shaki, F. Clarithromycin effectively enhances doxorubicin-induced cytotoxicity and apoptosis in MCF7 cells through dysregulation of autophagy. Adv. Med. Sci. 2020, 65, 235–243. [Google Scholar] [CrossRef]
- Bober, P.; Alexovic, M.; Tomkova, Z.; Kilik, R.; Sabo, J. RHOA and mDia1 Promotes Apoptosis of Breast Cancer Cells Via a High Dose of Doxorubicin Treatment. Open Life Sci. 2019, 14, 619–627. [Google Scholar] [CrossRef]
- Bates, P.J.; Reyes-Reyes, E.M.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1414–1428. [Google Scholar] [CrossRef] [PubMed]
- Alibolandi, M.; Ramezani, M.; Abnous, K.; Hadizadeh, F. AS1411 Aptamer-Decorated Biodegradable Polyethylene Glycol-Poly(lactic-co-glycolic acid) Nanopolymersomes for the Targeted Delivery of Gemcitabine to Non-Small Cell Lung Cancer In Vitro. J. Pharm. Sci. 2016, 105, 1741–1750. [Google Scholar] [CrossRef] [Green Version]
- Malik, M.T.; O’Toole, M.G.; Casson, L.K.; Thomas, S.D.; Bardi, G.T.; Reyes-Reyes, E.M.; Ng, C.K.; Kang, K.A.; Bates, P.J. AS1411-conjugated gold nanospheres and their potential for breast cancer therapy. Oncotarget 2015, 6, 22270–22281. [Google Scholar] [CrossRef] [Green Version]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 440. [Google Scholar] [CrossRef] [Green Version]
- Pereira, R.L.; Nascimento, I.C.; Santos, A.P.; Ogusuku, I.E.Y.; Lameu, C.; Mayer, G.; Ulrich, H. Aptamers: Novelty tools for cancer biology. Oncotarget 2018, 9, 26934–26953. [Google Scholar] [CrossRef] [Green Version]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Nieves-Neira, W.; Boon, C.; Pommier, Y.; Bonner, W.M. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J. Biol. Chem. 2000, 275, 9390–9395. [Google Scholar] [CrossRef] [Green Version]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, D.; Dong, F.; Wang, Y.Z.; Kayda, D.; Pledger, W.J. Regulation of cyclin E and p27kip during mitosis in BALB/c 3T3 cells. Cell Growth Differ. 1995, 6, 1199–1205. [Google Scholar] [PubMed]
- Frezza, V.; Pinto-Diez, C.; Fernandez, G.; Soto, M.; Martin, M.E.; Garcia-Sacristan, A.; Gonzalez, V.M. DNA aptamers targeting Leishmania infantum H3 protein as potential diagnostic tools. Anal. Chim. Acta 2020, 1107, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrión-Marchante, R.; Frezza, V.; Salgado-Figueroa, A.; Pérez-Morgado, M.I.; Martín, M.E.; González, V.M. DNA Aptamers against Vaccinia-Related Kinase (VRK) 1 Block Proliferation in MCF7 Breast Cancer Cells. Pharmaceuticals 2021, 14, 473. https://doi.org/10.3390/ph14050473
Carrión-Marchante R, Frezza V, Salgado-Figueroa A, Pérez-Morgado MI, Martín ME, González VM. DNA Aptamers against Vaccinia-Related Kinase (VRK) 1 Block Proliferation in MCF7 Breast Cancer Cells. Pharmaceuticals. 2021; 14(5):473. https://doi.org/10.3390/ph14050473
Chicago/Turabian StyleCarrión-Marchante, Rebeca, Valerio Frezza, Ana Salgado-Figueroa, M. Isabel Pérez-Morgado, M. Elena Martín, and Víctor M. González. 2021. "DNA Aptamers against Vaccinia-Related Kinase (VRK) 1 Block Proliferation in MCF7 Breast Cancer Cells" Pharmaceuticals 14, no. 5: 473. https://doi.org/10.3390/ph14050473
APA StyleCarrión-Marchante, R., Frezza, V., Salgado-Figueroa, A., Pérez-Morgado, M. I., Martín, M. E., & González, V. M. (2021). DNA Aptamers against Vaccinia-Related Kinase (VRK) 1 Block Proliferation in MCF7 Breast Cancer Cells. Pharmaceuticals, 14(5), 473. https://doi.org/10.3390/ph14050473