Mapping the Lipids of Skin Sebaceous Glands and Hair Follicles by High Spatial Resolution MALDI Imaging Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
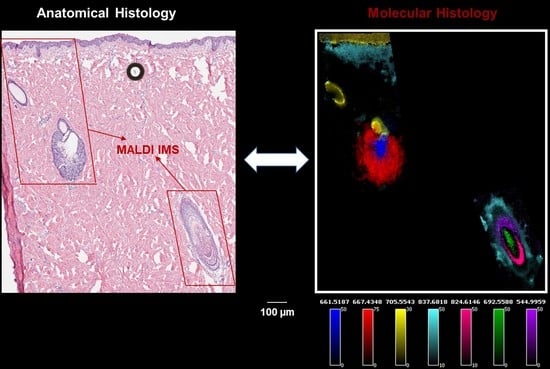
2.1. Identification of Lipids Associated with the Physiological Stages of the Sebaceous Glands (SG)
2.2. Identification of Lipids Associated with the Specific Regions of the Hair Follicles
3. Materials and Methods
3.1. Materials and Reagents
3.2. Tissue Sample Preparation
3.3. MALDI Imaging Mass Spectrometry
3.4. LC–MS Analysis
3.5. IMS and LC–MS Data Processing and Multivariate Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dilillo, M.; Heijs, B.; McDonnell, L.A. Mass spectrometry imaging: How will it affect clinical research in the future? Expert Rev. Proteom. 2018, 15, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Walch, A.; Rauser, S.; Deininger, S.O.; Hofler, H. MALDI imaging mass spectrometry for direct tissue analysis: A new frontier for molecular histology. Histochem. Cell Biol. 2008, 130, 421–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaurand, P.; Schwartz, S.A.; Billheimer, D.; Xu, B.J.; Crecelius, A.; Caprioli, R.M. Integrating histology and imaging mass spectrometry. Anal. Chem. 2004, 76, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
- Murillo, N.; Raoult, D. Skin microbiota: Overview and role in the skin diseases acne vulgaris and rosacea. Future Microbiol. 2013, 8, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Dahlhoff, M.; Camera, E.; Schäfer, M.; Emrich, D.; Riethmacher, D.; Foster, A.; Paus, R.; Schneider, M.R. Sebaceous lipids are essential for water repulsion, protection against UVB-induced apoptosis and ocular integrity in mice. Development 2016, 143, 1823–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigopoulos, D.; Stamatios, G.; Ioannides, D. Primary scarring alopecias. Alopecias-Pract. Eval. Manag. 2015, 47, 76–86. [Google Scholar]
- Schneider, M.R.; Paus, R. Deciphering the functions of the hair follicle infundibulum in skin physiology and disease. Cell Tissue Res. 2014, 358, 697–704. [Google Scholar] [CrossRef]
- Hunt, D.W.; Winters, G.C.; Brownsey, R.W.; Kulpa, J.E.; Gilliland, K.L.; Thiboutot, D.M.; Hofland, H.E. Inhibition of Sebum Production with the Acetyl Coenzyme A Carboxylase Inhibitor Olumacostat Glasaretil. J. Investig. Dermatol. 2017, 137, 1415–1423. [Google Scholar] [CrossRef] [Green Version]
- Chourasia, R.; Jain, S.K. Drug targeting through pilosebaceous route. Curr. Drug Targets 2009, 10, 950–967. [Google Scholar] [CrossRef]
- Patzelt, A.; Lademann, J. Drug delivery to hair follicles. Expert Opin. Drug Deliv. 2013, 10, 787–797. [Google Scholar] [CrossRef]
- Stenn, K.S.; Karnik, P. Lipids to the top of hair biology. J. Investig. Dermatol. 2010, 130, 1205–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, Y.; Tam, J.; Jalian, H.R.; Anderson, R.R.; Evans, C.L. Longitudinal, 3D in vivo imaging of sebaceous glands by coherent anti-stokes Raman scattering microscopy: Normal function and response to cryotherapy. J. Investig. Dermatol. 2015, 135, 39–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ford, S.J.; Bigliardi, P.L.; Sardella, T.C.; Urich, A.; Burton, N.C.; Kacprowicz, M.; Bigliardi, M.; Olivo, M.; Razansky, D. Structural and Functional Analysis of Intact Hair Follicles and Pilosebaceous Units by Volumetric Multispectral Optoacoustic Tomography. J. Investig. Dermatol. 2016, 136, 753–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trede, D.; Schiffler, S.; Becker, M.; Wirtz, S.; Steinhorst, K.; Strehlow, J.; Aichler, M.; Kobarg, J.H.; Oetjen, J.; Dyatlov, A.; et al. Exploring three-dimensional matrix-assisted laser desorption/ionization imaging mass spectrometry data: Three-dimensional spatial segmentation of mouse kidney. Anal. Chem. 2012, 84, 6079–6087. [Google Scholar] [CrossRef]
- Abirami, K.; Mayilvahanan, P. Performance Analysis of K-Means and Bisecting K-Means Algorithms in Weblog Data. Int. J. Emerg. Technol. Eng. Res. 2016, 4, 119–124. [Google Scholar]
- Wang, X.; Wang, X.; Liu, J.; Cai, T.; Guo, L.; Wang, S.; Wang, J.; Cao, Y.; Ge, J.; Jiang, Y.; et al. Hair Follicle and Sebaceous Gland De Novo Regeneration with Cultured Epidermal Stem Cells and Skin-Derived Precursors. Stem Cells Transl. Med. 2016, 5, 1695–1706. [Google Scholar] [CrossRef] [PubMed]
- Camera, E.; Ludovici, M.; Tortorella, S.; Sinagra, J.L.; Capitanio, B.; Goracci, L.; Picardo, M. Use of lipidomics to investigate sebum dysfunction in juvenile acne. J. Lipid Res. 2016, 57, 1051–1058. [Google Scholar] [CrossRef] [Green Version]
- Sjövall, P.; Skedung, L.; Gregoire, S.; Biganska, O.; Clément, F.; Luengo, G.S. Imaging the distribution of skin lipids and topically applied compounds in human skin using mass spectrometry. Sci. Rep. 2018, 8, 16683. [Google Scholar] [CrossRef] [Green Version]
- Engel, K.M.; Prabutzki, P.; Leopold, J.; Nimptsch, A.; Lemmnitzer, K.; Vos, D.N.; Hopf, C.; Schiller, J. A new update of MALDI-TOF mass spectrometry in lipid research. Prog. Lipid Res. 2022, 86, 101145. [Google Scholar] [CrossRef]
- Picardo, M.; Ottaviani, M.; Camera, E.; Mastrofrancesco, A. Sebaceous gland lipids. Dermatoendocrinol 2009, 1, 68–71. [Google Scholar] [CrossRef]
- Drake, D.R.; Brogden, K.A.; Dawson, D.V.; Wertz, P.W. Thematic review series: Skin lipids. Antimicrobial lipids at the skin surface. J. Lipid Res. 2008, 49, 4–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floettmann, E.; Lees, D.; Seeliger, F.; Jones, H.B. Pharmacological inhibition of DGAT1 induces sebaceous gland atrophy in mouse and dog skin while overt alopecia is restricted to the mouse. Toxicol. Pathol. 2015, 43, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Hart, P.J.; Francese, S.; Claude, E.; Woodroofe, M.N.; Clench, M.R. MALDI-MS imaging of lipids in ex vivo human skin. Anal. Bioanal. Chem. 2011, 401, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S. Integral hair lipid in human hair follicle. J. Dermatol. Sci. 2011, 64, 153–158. [Google Scholar] [CrossRef]
- Veijouye, S.J.; Abazar, Y.A.R.I.; Heidari, F.; Sajedi, N.; Moghani, F.G.; Nobakht, M. Bulge Region as a Putative Hair Follicle Stem Cells Niche: A Brief Review. Iran. J. Public Health 2017, 46, 1167–1175. [Google Scholar]
- Myung, P.; Ito, M. Dissecting the bulge in hair regeneration. J. Clin. Investig. 2012, 122, 448–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallégot, P.; Peteranderl, R.; Lechene, C. In-situ imaging mass spectrometry analysis of melanin granules in the human hair shaft. J. Investig. Dermatol. 2004, 122, 381–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt-Ullrich, R.; Paus, R. Molecular principles of hair follicle induction and morphogenesis. Bioessays 2005, 27, 247–261. [Google Scholar] [CrossRef]
- Mesler, A.L.; Veniaminova, N.A.; Lull, M.V.; Wong, S.Y. Hair Follicle Terminal Differentiation Is Orchestrated by Distinct Early and Late Matrix Progenitors. Cell Rep. 2017, 19, 809–821. [Google Scholar] [CrossRef]
- Lenoir, M.C.; Bernard, B.A.; Pautrat, G.; Darmon, M.; Shroot, B. Outer root sheath cells of human hair follicle are able to regenerate a fully differentiated epidermis in vitro. Dev. Biol. 1988, 130, 610–620. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knittelfelder, O.L.; Weberhofer, B.P.; Eichmann, T.O.; Kohlwein, S.D.; Rechberger, G.N. A versatile ultra-high performance LC-MS method for lipid profiling. J. Chromatogr. B 2014, 951–952, 119–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tortorella, S.; Tiberi, P.; Bowman, A.P.; Claes, B.S.; Scupakova, K.; Heeren, R.M.; Ellis, S.R.; Cruciani, G. LipostarMSI: Comprehensive, Vendor-Neutral Software for Visualization, Data Analysis, and Automated Molecular Identification in Mass Spectrometry Imaging. J. Am. Soc. Mass. Spectrom. 2020, 31, 155–163. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, L.A.; van Remoortere, A.; van Zeijl, R.J.; Deelder, A.M. Mass spectrometry image correlation: Quantifying colocalization. J. Proteome Res. 2008, 7, 3619–3627. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, F.; Groseclose, M.R.; Tortorella, S.; Cruciani, G.; Castellino, S. Mapping the Lipids of Skin Sebaceous Glands and Hair Follicles by High Spatial Resolution MALDI Imaging Mass Spectrometry. Pharmaceuticals 2022, 15, 411. https://doi.org/10.3390/ph15040411
Xie F, Groseclose MR, Tortorella S, Cruciani G, Castellino S. Mapping the Lipids of Skin Sebaceous Glands and Hair Follicles by High Spatial Resolution MALDI Imaging Mass Spectrometry. Pharmaceuticals. 2022; 15(4):411. https://doi.org/10.3390/ph15040411
Chicago/Turabian StyleXie, Fang, Mark Reid Groseclose, Sara Tortorella, Gabriele Cruciani, and Stephen Castellino. 2022. "Mapping the Lipids of Skin Sebaceous Glands and Hair Follicles by High Spatial Resolution MALDI Imaging Mass Spectrometry" Pharmaceuticals 15, no. 4: 411. https://doi.org/10.3390/ph15040411
APA StyleXie, F., Groseclose, M. R., Tortorella, S., Cruciani, G., & Castellino, S. (2022). Mapping the Lipids of Skin Sebaceous Glands and Hair Follicles by High Spatial Resolution MALDI Imaging Mass Spectrometry. Pharmaceuticals, 15(4), 411. https://doi.org/10.3390/ph15040411