Thymoquinone Enhances Apoptosis of K562 Chronic Myeloid Leukemia Cells through Hypomethylation of SHP-1 and Inhibition of JAK/STAT Signaling Pathway
Abstract
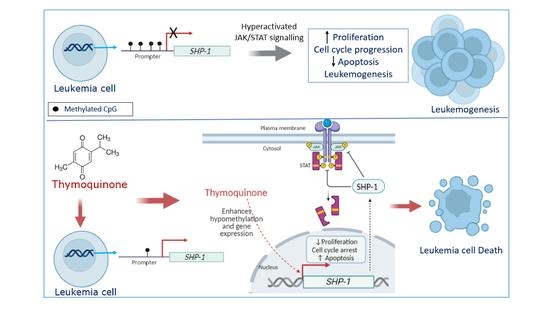
:1. Introduction
2. Results
2.1. Thymoquinone Enhances Apoptosis in K562 CML Cells
2.2. Thymoquinone Delays Cell Cycle Progression in K562 Cells
2.3. Thymoquinone Develops a Balanced EXPRESSION of the Genes That Regulate DNA Methylation in K562 CML Cells
2.4. Thymoquinone Causes Hypomethylation of the SHP-1 Promoter in K562 CML Cells
2.5. Thymoquinone Enhances SHP-1 Expression in K562 Cells
2.6. Thymoquinone Induces JAK/STAT Signaling Inhibition in K562 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Preparation of 5-Azacytidine and Thymoquinone Solutions
4.3. Analysis of Apoptosis by Flow Cytometer
4.4. Cell Cycle Analysis by PI Staining Assay
4.5. RT-qPCR for the Analysis of Gene Expression
4.6. Extraction and Bisulfite Treatment of DNA
4.7. Designing Primers for Pyrosequencing Analysis
4.8. Pyrosequencing Analysis
4.9. Preparation of Cellular Lysates
4.10. Evaluation of Protein Intensities by Jess-Simple Western Analysis
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Jamal, H.A.N.; Johan, M.F.; Jusoh, S.A.M.; Ismail, I.; Taib, W.R.W. Re-expression of bone marrow proteoglycan-2 by 5-azacytidine is associated with STAT3 inactivation and sensitivity response to imatinib in resistant CML cells. Asian Pac. J. Cancer Prev. 2018, 19, 1585. [Google Scholar] [PubMed]
- Al-Jamal, H.A.; Jusoh, S.A.M.; Yong, A.C.; Asan, J.M.; Hassan, R.; Johan, M.F. Silencing of suppressor of cytokine signaling-3 due to methylation results in phosphorylation of STAT3 in imatinib resistant BCR-ABL positive chronic myeloid leukemia cells. Asian Pac. J. Cancer Prev. 2014, 15, 4555–4561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Liu, X.; Guo, X.; Liu, X.; Luo, J. DNA methyltransferase 1 mediated aberrant methylation and silencing of SHP-1 gene in chronic myelogenous leukemia cells. Leuk. Res. 2017, 58, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Koschmieder, S.; Vetrie, D. Epigenetic dysregulation in chronic myeloid leukaemia: A myriad of mechanisms and therapeutic options. Semin. Cancer Biol. 2018, 51, 180–197. [Google Scholar] [CrossRef]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef] [Green Version]
- Luo, P.; Jing, W.; Yi, K.; Wu, S.; Zhou, F. Wilms’ tumor 1 gene in hematopoietic malignancies: Clinical implications and future directions. Leuk. Lymphoma 2020, 61, 2059–2067. [Google Scholar] [CrossRef]
- Al-Jamal, H.A.N.; Taib, W.R.W.; Jusoh, S.A.M.; Sudin, A.; Johan, M.F. Azacytidine Enhances Sensitivity Response to Imatinib in BCR/ABL positive CML Cell Line. J. Biomed. Sci. 2018, 2, 20–22. [Google Scholar]
- Wu, M.; Song, D.; Li, H.; Yang, Y.; Ma, X.; Deng, S.; Ren, C.; Shu, X. Negative regulators of STAT3 signaling pathway in cancers. Cancer Manag. Res. 2019, 11, 4957. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Hong, Q.; Shen, Y.; Xu, Y.; Zhu, H.; Li, Y.; Xu, C.; Ouyang, G.; Duan, S. The diagnostic value of DNA methylation in leukemia: A systematic review and meta-analysis. PLoS ONE 2014, 9, e96822. [Google Scholar] [CrossRef]
- Groner, B.; von Manstein, V. Jak Stat signaling and cancer: Opportunities, benefits and side effects of targeted inhibition. Mol. Cell. Endocrinol. 2017, 451, 1–14. [Google Scholar] [CrossRef]
- Xiong, H.; Chen, Z.F.; Liang, Q.C.; Du, W.; Chen, H.M.; Su, W.Y.; Chen, G.Q.; Han, Z.G.; Fang, J.Y. Inhibition of DNA methyltransferase induces G2 cell cycle arrest and apoptosis in human colorectal cancer cells via inhibition of JAK2/STAT3/STAT5 signalling. J. Cell. Mol. Med. 2009, 13, 3668–3679. [Google Scholar] [CrossRef]
- Agrawal, K.; Das, V.; Vyas, P.; Hajdúch, M. Nucleosidic DNA demethylating epigenetic drugs—A comprehensive review from discovery to clinic. Pharmacol. Ther. 2018, 188, 45–79. [Google Scholar] [CrossRef]
- Yan, F.; Shen, N.; Pang, J.; Molina, J.R.; Yang, P.; Liu, S. The DNA methyltransferase DNMT1 and tyrosine-protein kinase KIT cooperatively promote resistance to 5-Aza-2′-deoxycytidine (decitabine) and midostaurin (PKC412) in lung cancer cells. J. Biol. Chem. 2015, 290, 18480–18494. [Google Scholar] [CrossRef] [Green Version]
- Al-Saraireh, Y.M.; Youssef, A.M.; Alsarayreh, A.; Al Hujran, T.A.; Al-Sarayreh, S.; Al-Shuneigat, J.M.; Alrawashdeh, H.M. Phytochemical and anti-cancer properties of Euphorbia hierosolymitana Boiss. crude extracts. J. Pharm. Pharmacogn. Res. 2021, 9, 13–23. [Google Scholar] [CrossRef]
- Al-saraireh, Y.M.; Youssef, A.M.; Alshammari, F.O.; Al-Sarayreh, S.A.; Al-Shuneigat, J.M.; Alrawashdeh, H.M.; Mahgoub, S.S. Phytochemical characterization and anti-cancer properties of extract of Ephedra foeminea (Ephedraceae) aerial parts. Trop. J. Pharm. Res. 2021, 20, 1675–1681. [Google Scholar] [CrossRef]
- Youssef, A.; El-Swaify, Z.; Al-saraireh, Y.; Dalain, S. Cytotoxic activity of methanol extract of Cynanchumacutum L. seeds on human cancer cell lines. Latin Am. J. Pharm. 2018, 37, 1997–2003. [Google Scholar]
- Youssef, A.M.; El-Swaify, Z.S.; Al-Saraireh, Y.; Al-Dalain, S. Anticancer effect of different extracts of Cynanchum acutum L. seeds on cancer cell lines. Pharmacogn. Mag. 2019, 15, 261–266. [Google Scholar] [CrossRef]
- Shankar, E.; Kanwal, R.; Candamo, M.; Gupta, S. Dietary phytochemicals as epigenetic modifiers in cancer: Promise and challenges. Semin. Cancer Biol. 2016, 40–41, 82–99. [Google Scholar] [CrossRef] [Green Version]
- Abdullah, O.; Omran, Z.; Hosawi, S.; Hamiche, A.; Bronner, C.; Alhosin, M. Thymoquinone is a multitarget single epidrug that inhibits the UHRF1 protein complex. Genes 2021, 12, 622. [Google Scholar] [CrossRef]
- Li, J.; Khan, M.; Wei, C.; Cheng, J.; Chen, H.; Yang, L.; Ijaz, I.; Fu, J. Thymoquinone inhibits the migration and invasive characteristics of cervical cancer cells SiHa and CaSki in vitro by targeting epithelial to mesenchymal transition associated transcription factors Twist1 and Zeb1. Molecules 2017, 22, 2105. [Google Scholar] [CrossRef] [Green Version]
- Khazaei, M.; Pazhouhi, M. Temozolomide-mediated apoptotic death is improved by thymoquinone in U87MG cell line. Cancer Investig. 2017, 35, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Alaufi, O.M.; Noorwali, A.; Zahran, F.; Al-Abd, A.M.; Al-Attas, S. Cytotoxicity of thymoquinone alone or in combination with cisplatin (CDDP) against oral squamous cell carcinoma in vitro. Sci. Rep. 2017, 7, 13131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Far, A.H.; Salaheldin, T.A.; Godugu, K.; Darwish, N.H.; Mousa, S.A. Thymoquinone and its nanoformulation attenuate colorectal and breast cancers and alleviate doxorubicin-induced cardiotoxicity. Nanomedicine 2021, 6, 1457–1469. [Google Scholar] [CrossRef] [PubMed]
- Soltani, A.; Pourgheysari, B.; Shirzad, H.; Sourani, Z. Antiproliferative and apoptosis-inducing activities of thymoquinone in lymphoblastic leukemia cell line. Indian J. Hematol. Blood Transfus. 2017, 33, 516–524. [Google Scholar] [CrossRef]
- Houssein, M.; Fatfat, M.; Habli, Z.; Ghazal, N.; Moodad, S.; Khalife, H.; Khalil, M.; Gali-Muhtasib, H. Thymoquinone synergizes with arsenic and interferon alpha to target human T-cell leukemia/lymphoma. Life Sci. 2020, 251, 117639. [Google Scholar] [CrossRef]
- AlGhamdi, A.A.; Mohammed, M.R.S.; Zamzami, M.A.; Al-Malki, A.L.; Qari, M.H.; Khan, M.I.; Choudhry, H. Untargeted metabolomics identifies key metabolic pathways altered by thymoquinone in leukemic cancer cells. Nutrients 2020, 12, 1792. [Google Scholar] [CrossRef]
- Pang, J.; Shen, N.; Yan, F.; Zhao, N.; Dou, L.; Wu, L.-C.; Seiler, C.L.; Yu, L.; Yang, K.; Bachanova, V. Thymoquinone exerts potent growth-suppressive activity on leukemia through DNA hypermethylation reversal in leukemia cells. Oncotarget 2017, 8, 34453. [Google Scholar] [CrossRef] [Green Version]
- Shanmugam, M.K.; Arfuso, F.; Kumar, A.P.; Wang, L.; Goh, B.C.; Ahn, K.S.; Bishayee, A.; Sethi, G. Modulation of diverse oncogenic transcription factors by thymoquinone, an essential oil compound isolated from the seeds of Nigella sativa Linn. Pharmacol. Res. 2018, 129, 357–364. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, Y.; Huang, S.; Wang, G.; Han, R.; Lei, F.; Luo, A.; Jing, X.; Zhao, L.; Gu, S. Thymoquinone inhibits the metastasis of renal cell cancer cells by inducing autophagy via AMPK/mTOR signaling pathway. Cancer Sci. 2018, 109, 3865–3873. [Google Scholar] [CrossRef]
- Liu, K.; Wu, Z.; Chu, J.; Yang, L.; Wang, N. Promoter methylation and expression of SOCS3 affect the clinical outcome of pediatric acute lymphoblastic leukemia by JAK/STAT pathway. Biomed. Pharmacother. 2019, 115, 108913. [Google Scholar] [CrossRef]
- Fatfat, M.; Fakhoury, I.; Habli, Z.; Mismar, R.; Gali-Muhtasib, H. Thymoquinone enhances the anticancer activity of doxorubicin against adult T-cell leukemia in vitro and in vivo through ROS-dependent mechanisms. Life Sci. 2019, 232, 116628. [Google Scholar] [CrossRef]
- Al-Rawashde, F.A.; Taib, W.R.W.; Ismail, I.; Johan, M.F.; Al-Wajeeh, A.S.; Al-Jamal, H.A.N. Thymoquinone Induces downregulation of BCR-ABL/JAK/STAT pathway and apoptosis in K562 leukemia cells. Asian Pac. J. Cancer Prev. 2021, 22, 3959. [Google Scholar] [CrossRef]
- Hoang, N.M.; Rui, L. DNA methyltransferases in hematological malignancies. J Genet. Genom. 2020, 47, 361–372. [Google Scholar] [CrossRef]
- Wang, Y.; Weng, W.-J.; Zhou, D.-H.; Fang, J.-P.; Mishra, S.; Chai, L.; Xu, L.-H. Wilms tumor 1 mutations are independent poor prognostic factors in pediatric acute myeloid leukemia. Front. Oncol. 2021, 11, 632094. [Google Scholar] [CrossRef]
- Rampal, R.; Figueroa, M.E. Wilms tumor 1 mutations in the pathogenesis of acute myeloid leukemia. Haematologica 2016, 101, 672. [Google Scholar] [CrossRef] [Green Version]
- Al-Rawashde, F.A.; Johan, M.F.; Taib, W.R.W.; Ismail, I.; Johari, S.A.T.T.; Almajali, B.; Al-Wajeeh, A.S.; Nazari Vishkaei, M.; Al-Jamal, H.A.N. Thymoquinone Inhibits Growth of Acute Myeloid Leukemia Cells through Reversal SHP-1 and SOCS-3 Hypermethylation: In Vitro and In Silico Evaluation. Pharmaceuticals 2021, 14, 1287. [Google Scholar] [CrossRef]
- Qadi, S.A.; Hassan, M.A.; Sheikh, R.A.; Baothman, O.A.; Zamzami, M.A.; Choudhry, H.; Al-Malki, A.L.; Albukhari, A.; Alhosin, M. Thymoquinone-induced reactivation of tumor suppressor genes in cancer cells involves epigenetic mechanisms. Epigenetics Insights 2019, 12, 2516865719839011. [Google Scholar] [CrossRef]
- Alhosin, M.; Razvi, S.S.I.; Sheikh, R.A.; Khan, J.A.; Zamzami, M.A.; Choudhry, H. Thymoquinone and Difluoromethylornithine (DFMO) Synergistically Induce Apoptosis of Human Acute T Lymphoblastic Leukemia Jurkat Cells Through the Modulation of Epigenetic Pathways. Technol. Cancer Res. Treat. 2020, 19, 1533033820947489. [Google Scholar] [CrossRef]
- Amartey, J.; Gapper, S.; Hussein, N.; Morris, K.; Withycombe, C.E. Nigella sativa Extract and Thymoquinone Regulate Inflammatory Cytokine and TET-2 Expression in Endothelial Cells. Artery Res. 2019, 25, 157–163. [Google Scholar] [CrossRef] [Green Version]
- Musalli, M.G.; Hassan, M.A.; Sheikh, R.A.; Kalantan, A.A.; Halwani, M.A.; Zeyadi, M.; Hosawi, S.; Alhosin, M. Thymoquinone induces cell proliferation inhibition and apoptosis in acute myeloid leukemia cells: Role of apoptosis-related WT1 and BCL2 genes. Eur. J. Cell Biol. 2019, 1, 2–9. [Google Scholar] [CrossRef]
- Toska, E.; Roberts, S.G. Mechanisms of transcriptional regulation by WT1 (Wilms’ tumour 1). Biochem. J. 2014, 461, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Goel, H.; Rahul, E.; Gupta, A.K.; Meena, J.P.; Chopra, A.; Ranjan, A.; Hussain, S.; Rath, G.; Tanwar, P. Molecular update on biology of Wilms Tumor 1 gene and its applications in acute myeloid leukemia. Am. J. Blood Res. 2020, 10, 151. [Google Scholar] [PubMed]
- Uhm, K.-O.; Lee, E.S.; Lee, Y.M.; Park, J.S.; Kim, S.J.; Kim, B.S.; Kim, H.S.; Park, S.-H. Differential methylation pattern of ID4, SFRP1, and SHP1 between acute myeloid leukemia and chronic myeloid leukemia. J. Korean Med. Sci. 2009, 24, 493–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.A.; Tania, M.; Wei, C.; Mei, Z.; Fu, S.; Cheng, J.; Xu, J.; Fu, J. Thymoquinone inhibits cancer metastasis by downregulating TWIST1 expression to reduce epithelial to mesenchymal transition. Oncotarget 2015, 6, 19580. [Google Scholar] [CrossRef] [Green Version]
- Asmaa, M.J.S.; Al-Jamal, H.A.; Hussein, A.R.; Yahaya, B.H.; Hassan, R.; Hussain, F.A.; Shamsuddin, S.; Johan, M.F. Transcriptomic profiles of MV4-11 and kasumi 1 acute myeloid leukemia cell lines modulated by epigenetic modifiers trichostatin A and 5-Azacytidine. Int. J. Hematol.-Oncol. Stem Cell Res. 2020, 14, 72. [Google Scholar] [CrossRef]
- Cai, H.; Qin, X.; Yang, C. Dehydrocostus lactone suppresses proliferation of human chronic myeloid leukemia cells through Bcr/Abl-JAK/STAT signaling pathways. J. Cell. Biochem. 2017, 118, 3381–3390. [Google Scholar] [CrossRef]
- Badr, G.; Mohany, M.; Abu-Tarboush, F. Thymoquinone decreases F-actin polymerization and the proliferation of human multiple myeloma cells by suppressing STAT3 phosphorylation and Bcl2/Bcl-XL expression. Lipids Health Dis. 2011, 10, 236. [Google Scholar] [CrossRef] [Green Version]
- Al-Rawashde, F.A.; Al-Wajeeh, A.S.; Vishkaei, M.N.; Saad, H.K.M.; Johan, M.F.; Taib, W.R.W.; Ismail, I.; Al-Jamal, H.A.N. Thymoquinone Inhibits JAK/STAT and PI3K/Akt/mTOR Signaling Pathways in MV4-11 and K562 Myeloid Leukemia Cells. Pharmaceuticals 2022, 15, 1123. [Google Scholar] [CrossRef]
- Almajali, B.; Johan, M.F.; Al-Wajeeh, A.S.; Wan Taib, W.R.; Ismail, I.; Alhawamdeh, M.; Al-Tawarah, N.M.; Ibrahim, W.N.; Al-Rawashde, F.A.; Al-Jamal, H.A.N. Gene Expression Profiling and Protein Analysis Reveal Suppression of the C-Myc Oncogene and Inhibition JAK/STAT and PI3K/AKT/mTOR Signaling by Thymoquinone in Acute Myeloid Leukemia Cells. Pharmaceuticals 2022, 15, 307. [Google Scholar] [CrossRef]
- Raut, P.K.; Lee, H.S.; Joo, S.H.; Chun, K.-S. Thymoquinone induces oxidative stress-mediated apoptosis through downregulation of Jak2/STAT3 signaling pathway in human melanoma cells. Food Chem. Toxicol. 2021, 157, 112604. [Google Scholar] [CrossRef]
- Hatiboglu, M.A.; Kocyigit, A.; Guler, E.M.; Akdur, K.; Nalli, A.; Karatas, E.; Tuzgen, S. Thymoquinone induces apoptosis in B16-F10 melanoma cell through inhibition of p-STAT3 and inhibits tumor growth in a murine intracerebral melanoma model. World Neurosurg. 2018, 114, e182–e190. [Google Scholar] [CrossRef]
- Ranjbari, A.; Heidarian, E.; Ghatreh-Samani, K. Effects of thymoquinone on IL-6 Gene expression and some cellular signaling pathways in prostate cancer PC3 cells. Jundishapur J. Nat. Pharm. Prod. 2017, 12, e63753. [Google Scholar] [CrossRef]
- Ibrahim, A.; Alhosin, M.; Papin, C.; Ouararhni, K.; Omran, Z.; Zamzami, M.A.; Al-Malki, A.L.; Choudhry, H.; Mély, Y.; Hamiche, A. Thymoquinone challenges UHRF1 to commit auto-ubiquitination: A key event for apoptosis induction in cancer cells. Oncotarget 2018, 9, 28599. [Google Scholar] [CrossRef] [Green Version]
- Khalife, R.; Stephany, E.-H.; Tarras, O.; Hodroj, M.H.; Rizk, S. Antiproliferative and proapoptotic effects of topotecan in combination with thymoquinone on acute myelogenous leukemia. Clin. Lymphoma Myeloma Leuk. 2014, 14, S46–S55. [Google Scholar] [CrossRef]
- Motaghed, M. Cytotoxic, cytostatic and anti-estrogenic effect of Thymoquinone on estrogen receptor-positive breast cancer MCF7 cell line. Am. J. Life Sci. 2015, 3, 7–14. [Google Scholar] [CrossRef] [Green Version]
- Kielkopf, C.L.; Bauer, W.; Urbatsch, I.L. Bradford assay for determining protein concentration. Cold Spring Harb. Protoc. 2020, 2020, 102269. [Google Scholar] [CrossRef]





| CpG.1 | CpG.2 | CpG.3 | CpG.4 | Mean ± Standard Deviation | |
|---|---|---|---|---|---|
| Untreated-K562 cells | 96 | 91 | 93 | 88 | 92.0 ± 22.8 |
| TQ-treated K562 cells | 89 | 83 | 70 | 83 | 81.2 ± 11.0 |
| 5-Aza-treated K562 cells | 70 | 58 | 68 | 63 | 64.7 ± 12.1 |
| Unmethylated DNA control | 6 | 0 | 0 | 0 | 1.5 ± 0.4 |
| Methylated DNA control | 95 | 88 | 100 | 84 | 91.8 ± 15.7 |
| Unmethylated Bisufite-unconverted DNA control | 0 | 0 | 0 | 0 | 0 |
| Proteins | M.Ws (kDa) | Proteins’ Intensities Medians (IqR) | p-Value a | |
|---|---|---|---|---|
| Untreated K562 Cells | TQ-treated K562 Cells | |||
| STAT3 | 80–90 | 3,891,993 (312,350) | 3,153,914 (262,434 | <0.05 |
| p-STAT3 | 95 | 2,559,531 (254,110) | 1,576,590 (131,188) | <0.001 |
| STAT5 | 91 | 8,924,147 (295,549) | 4,762,098 (192,331) | <0.001 |
| p-STAT5 | 92 | 1,648,065 (170,566) | 1,221,741 (154,917) | <0.05 |
| JAK2 | 130 | 21,887,799 (501,710) | 20,385,796 (423,651) | <0.05 |
| p-JAK2 | 130 | 1,225,525 (103,221) | 601,966 (91,423) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Rawashde, F.A.; Al-Sanabra, O.M.; Alqaraleh, M.; Jaradat, A.Q.; Al-Wajeeh, A.S.; Johan, M.F.; Wan Taib, W.R.; Ismail, I.; Al-Jamal, H.A.N. Thymoquinone Enhances Apoptosis of K562 Chronic Myeloid Leukemia Cells through Hypomethylation of SHP-1 and Inhibition of JAK/STAT Signaling Pathway. Pharmaceuticals 2023, 16, 884. https://doi.org/10.3390/ph16060884
Al-Rawashde FA, Al-Sanabra OM, Alqaraleh M, Jaradat AQ, Al-Wajeeh AS, Johan MF, Wan Taib WR, Ismail I, Al-Jamal HAN. Thymoquinone Enhances Apoptosis of K562 Chronic Myeloid Leukemia Cells through Hypomethylation of SHP-1 and Inhibition of JAK/STAT Signaling Pathway. Pharmaceuticals. 2023; 16(6):884. https://doi.org/10.3390/ph16060884
Chicago/Turabian StyleAl-Rawashde, Futoon Abedrabbu, Ola M. Al-Sanabra, Moath Alqaraleh, Ahmad Q. Jaradat, Abdullah Saleh Al-Wajeeh, Muhammad Farid Johan, Wan Rohani Wan Taib, Imilia Ismail, and Hamid Ali Nagi Al-Jamal. 2023. "Thymoquinone Enhances Apoptosis of K562 Chronic Myeloid Leukemia Cells through Hypomethylation of SHP-1 and Inhibition of JAK/STAT Signaling Pathway" Pharmaceuticals 16, no. 6: 884. https://doi.org/10.3390/ph16060884
APA StyleAl-Rawashde, F. A., Al-Sanabra, O. M., Alqaraleh, M., Jaradat, A. Q., Al-Wajeeh, A. S., Johan, M. F., Wan Taib, W. R., Ismail, I., & Al-Jamal, H. A. N. (2023). Thymoquinone Enhances Apoptosis of K562 Chronic Myeloid Leukemia Cells through Hypomethylation of SHP-1 and Inhibition of JAK/STAT Signaling Pathway. Pharmaceuticals, 16(6), 884. https://doi.org/10.3390/ph16060884