CYP2D6 Genotype and Pharmacovigilance Impact on Autism Spectrum Disorder: A Naturalistic Study with Extreme Phenotype Analysis
Abstract
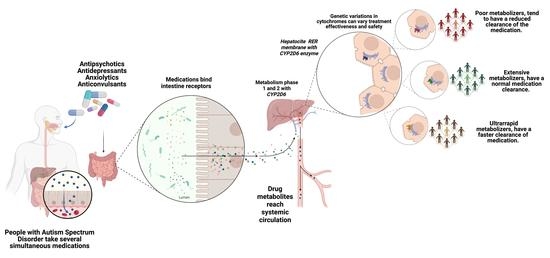
:1. Introduction
2. Results
2.1. Demographic and Clinical Data
2.2. CYP2D6 Phenotype Influence
2.3. Pharmacology Variables
2.4. CYP2D6 Phenotype Influence
2.5. Case Reports Series
3. Discussion
4. Method
4.1. Study Design and Ethics
4.2. Participants
4.3. Procedure
4.4. Data Collection
4.5. Comorbidities and AEs
4.6. CYP2D6 Genotyping and Phenotyping
4.7. Drug Reports: CYP2D6 Phenotype Applied to Clinical Cases
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jobski, K.; Höfer, J.; Hoffmann, F.; Bachmann, C. Use of psychotropic drugs in patients with autism spectrum disorders: A systematic review. Acta Psychiatr. Scand. 2017, 135, 8–28. [Google Scholar] [CrossRef]
- Espadas, C.; Ballester, P.; Londoño, A.C.; Almenara, S.; Aguilar, V.; Belda, C.; Pérez, E.; Peiró, A.M. Multimorbidity and psychotropic polypharmacy among participants with autism spectrum disorder with intellectual disability. Psychiatry Res. 2020, 292, 113321. [Google Scholar] [CrossRef]
- Joshi, G.; Petty, C.; Wozniak, J.; Henin, A.; Fried, R.; Galdo, M.; Kotarski, M.; Walls, S.; Biederman, J. The heavy burden of psychiatric comorbidity in youth with autism spectrum disorders: A large comparative study of a psychiatrically referred population. J. Autism Dev. Disord. 2010, 40, 1361–1370. [Google Scholar] [CrossRef]
- Accordino, R.E.; Kidd, C.; Politte, L.C.; Henry, C.A.; McDougle, C.J. Psychopharmacological interventions in autism spectrum disorder. Expert Opin. Pharmacother. 2016, 17, 937–952. [Google Scholar] [CrossRef] [PubMed]
- Houghton, R.; Ong, R.C.; Bolognani, F. Psychiatric comorbidities and use of psychotropic medications in people with autism spectrum disorder in the United States. Autism Res. 2017, 10, 2037–2047. [Google Scholar] [CrossRef]
- Rigles, B. The Relationship Between Adverse Childhood Events, Resiliency and Health Among Children with Autism. J. Autism Dev. Disord. 2017, 47, 187–202. [Google Scholar] [CrossRef]
- Gerlach, M.; Egberts, K.; Dang, S.Y.; Plener, P.; Taurines, R.; Mehler-Wex, C.; Romanos, M. Therapeutic drug monitoring as a measure of proactive pharmacovigilance in child and adolescent psychiatry. Expert Opin. Drug Saf. 2016, 15, 1477–1482. [Google Scholar] [CrossRef]
- Crews, K.R.; Gaedigk, A.; Dunnenberger, H.M.; Leeder, J.S.; Klein, T.E.; Caudle, K.E.; Haidar, C.E.; Shen, D.D.; Callaghan, J.T.; Sadhasivam, S.; et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. 2014, 95, 376–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buck, T.R.; Viskochil, J.; Farley, M.; Coon, H.; McMahon, W.M.; Morgan, J.; Bilder, D.A. Psychiatric comorbidity and medication use in adults with autism spectrum disorder. J. Autism Dev. Disord. 2014, 44, 3063–3071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartwright, A.L.; Wilby, K.J.; Corrigan, S.; Ensom, M.H.H. Pharmacogenetics of risperidone: A systematic review of the clinical effects of CYP2D6 polymorphisms. Ann. Pharmacother. 2013, 47, 350–360. [Google Scholar] [CrossRef]
- Vanwong, N.; Ngamsamut, N.; Hongkaew, Y.; Nuntamool, N.; Puangpetch, A.; Chamnanphon, M.; Sinrachatanant, A.; Limsila, P.; Sukasem, C. Detection of CYP2D6 polymorphism using Luminex xTAG technology in autism spectrum disorder: CYP2D6 activity score and its association with risperidone levels. Drug Metab. Pharmacokinet. 2016, 31, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Vanwong, N.; Ngamsamut, N.; Medhasi, S.; Puangpetch, A.; Chamnanphon, M.; Tan-Kam, T.; Hongkaew, Y.; Limsila, P.; Sukasem, C. Impact of CYP2D6 Polymorphism on Steady-State Plasma Levels of Risperidone and 9-Hydroxyrisperidone in Thai Children and Adolescents with Autism Spectrum Disorder. J. Child Adolesc. Psychopharmacol. 2017, 27, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Thümmler, S.; Dor, E.; David, R.; Leali, G.; Battista, M.; David, A.; Askenazy, F.; Verstuyft, C. Pharmacoresistant Severe Mental Health Disorders in Children and Adolescents: Functional Abnormalities of Cytochrome P450 2D6. Front. Psychiatry 2018, 9, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samer, C.F.; Lorenzini, K.I.; Rollason, V.; Daali, Y.; Desmeules, J.A. Applications of CYP450 testing in the clinical setting. Mol. Diagn. Ther. 2013, 17, 165–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fijal, B.A.; Guo, Y.; Li, S.G.; Ahl, J.; Goto, T.; Tanaka, Y.; Nisenbaum, L.K.; Upadhyaya, H.P. CYP2D6 predicted metabolizer status and safety in adult patients with attention-deficit hyperactivity disorder participating in a large placebo-controlled atomoxetine maintenance of response clinical trial. J. Clin. Pharmacol. 2015, 55, 1167–1174. [Google Scholar] [CrossRef]
- Candiotti, K.A.; Yang, Z.; Rodriguez, Y.; Crescimone, A.; Sanchez, G.C.; Takacs, P.; Medina, C.; Zhang, Y.; Liu, H.; Gitlin, M.C. The impact of CYP2D6 genetic polymorphisms on postoperative morphine consumption. Pain Med. 2009, 10, 799–805. [Google Scholar] [CrossRef]
- Lopes, G.S.; Bielinski, S.J.; Moyer, A.M.; Black Lii, J.L.; Jacobson, D.J.; Jiang, R.; Larson, N.B.; St Sauver, J.L. Sex Differences in Associations Between CYP2D6 Phenotypes and Response to Opioid Analgesics. Pharm. Pers. Med. 2020, 13, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Kirchheiner, J.; Henckel, H.B.; Meineke, I.; Roots, I.; Brockmöller, J. Impact of the CYP2D6 ultrarapid metabolizer genotype on mirtazapine pharmacokinetics and adverse events in healthy volunteers. J. Clin. Psychopharmacol. 2004, 24, 647–652. [Google Scholar] [CrossRef]
- Anderson, G.D. Gender differences in pharmacological response. Int. Rev. Neurobiol. 2008, 83, 1–10. [Google Scholar] [CrossRef]
- Haufroid, V.; Hantson, P. CYP2D6 genetic polymorphisms and their relevance for poisoning due to amfetamines, opioid analgesics and antidepressants. Clin. Toxicol. 2015, 53, 501–510. [Google Scholar] [CrossRef]
- Lötsch, J.; Skarke, C.; Liefhold, J.; Geisslinger, G. Genetic predictors of the clinical response to opioid analgesics: Clinical utility and future perspectives. Clin. Pharmacokinet. 2004, 43, 983–1013. [Google Scholar] [CrossRef] [PubMed]
- Zahari, Z.; Ismail, R. Influence of Cytochrome P450, Family 2, Subfamily D, Polypeptide 6 (CYP2D6) polymorphisms on pain sensitivity and clinical response to weak opioid analgesics. Drug Metab. Pharmacokinet. 2014, 29, 29–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vohra, R.; Madhavan, S.; Sambamoorthi, U. Comorbidity prevalence, healthcare utilization, and expenditures of Medicaid enrolled adults with autism spectrum disorders. Autism Int. J. Res. Pract. 2017, 21, 995–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correll, C.U.; Penzner, J.B.; Parikh, U.H.; Mughal, T.; Javed, T.; Carbon, M.; Malhotra, A.K. Recognizing and monitoring adverse events of second-generation antipsychotics in children and adolescents. Child Adolesc. Psychiatr. Clin. N. Am. 2006, 15, 177–206. [Google Scholar] [CrossRef]
- Correll, C.U.; Manu, P.; Olshanskiy, V.; Napolitano, B.; Kane, J.M.; Malhotra, A.K. Cardiometabolic risk of second-generation antipsychotic medications during first-time use in children and adolescents. JAMA 2009, 302, 1765–1773. [Google Scholar] [CrossRef] [Green Version]
- Arellano, A.L.; Martin-Subero, M.; Monerris, M.; Llerena, A.; Farré, M.; Montané, E. Multiple adverse drug reactions and genetic polymorphism testing: A case report with negative result. Medicine 2017, 96, e8505. [Google Scholar] [CrossRef]
- Belmonte, C.; Ochoa, D.; Román, M.; Saiz-Rodríguez, M.; Wojnicz, A.; Gómez-Sánchez, C.I.; Martín-Vílchez, S.; Abad-Santos, F. Influence of CYP2D6, CYP3A4, CYP3A5 and ABCB1 Polymorphisms on Pharmacokinetics and Safety of Aripiprazole in Healthy Volunteers. Basic Clin. Pharmacol. Toxicol. 2018, 122, 596–605. [Google Scholar] [CrossRef] [Green Version]
- Bahar, M.A.; Wang, Y.; Bos, J.H.J.; Wilffert, B.; Hak, E. Discontinuation and dose adjustment of metoprolol after metoprolol-paroxetine/fluoxetine co-prescription in Dutch elderly. Pharmacoepidemiol. Drug Saf. 2018, 27, 621–629. [Google Scholar] [CrossRef] [Green Version]
- Ching, H.; Pringsheim, T. Aripiprazole for autism spectrum disorders (ASD). Cochrane Database Syst. Rev. 2012, 6, 621–629. [Google Scholar] [CrossRef] [Green Version]
- Posey, D.J.; Stigler, K.A.; Erickson, C.A.; McDougle, C.J. Antipsychotics in the treatment of autism. J. Clin. Investig. 2008, 118, 6–14. [Google Scholar] [CrossRef]
- Puangpetch, A.; Vanwong, N.; Nuntamool, N.; Hongkaew, Y.; Chamnanphon, M.; Sukasem, C. CYP2D6 polymorphisms and their influence on risperidone treatment. Pharm. Pers. Med. 2016, 9, 131–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, B.; Gastwirth, J.L. A method of assessing the sensitivity of the Cochran-Mantel-Haenszel test to an unobserved confounder. Philos. Trans. Ser. A Math. Phys. Eng. Sci. 2008, 366, 2377–2388. [Google Scholar] [CrossRef] [PubMed]
- Caudle, K.E.; Sangkuhl, K.; Whirl-Carrillo, M.; Swen, J.J.; Haidar, C.E.; Klein, T.E.; Gammal, R.S.; Relling, M.V.; Scott, S.A.; Hertz, D.L.; et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmaco-genetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 2020, 13, 116–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaedigk, A.; Sangkuhl, K.; Whirl-Carrillo, M.; Klein, T.; Steven Leeder, J. Prediction of CYP2D6 phenotype from genotype across world populations. Genet. Med. 2017, 19, 69–76. [Google Scholar] [CrossRef] [Green Version]
- LLerena, A.; Naranjo, M.E.G.; Rodrigues-Soares, F.; Penas-LLedó, E.M.; Fariñas, H.; Tarazona-Santos, E. Interethnic variability of CYP2D6 alleles and of predicted and measured metabolic phenotypes across world populations. Expert Opin. Drug Metab. Toxicol. 2014, 10, 1569–1583. [Google Scholar] [CrossRef] [PubMed]
- Naranjo, M.E.G.; De Andrés, F.; Delgado, A.; Cobaleda, J.; Peñas-Lledó, E.M.; Llerena, A. High frequency of CYP2D6 ultrarapid metabolizers in Spain: Controversy about their misclassification in worldwide population studies. Pharm. J. 2016, 16, 485–490. [Google Scholar] [CrossRef]



| Population | ASD (n = 83) * | ASD-CYP2D6 (n = 71) | PM (n = 5) | IM (n = 31) | NM (n = 32) | UM (n = 3) |
|---|---|---|---|---|---|---|
| Age (mean (SD) years) | 30 (10) | 30 (10) | 27 (9) | 29 (10) | 30 (10) | 33(10) |
| Sex (% male) | 86% | 82% | 80% | 84% | 81% | 100% |
| Intellectual disability in HER ** (IQ < 70) | 20% | 20% | 20% | 22.5% | 17% | 33% |
| Poor Metabolizers (n = 5, 7%) | Comorbidities (n = 222) |
| Nervous System (10/16) | Anxiety (2/10) |
| Depression (1/10) | |
| Epilepsy (3/10) | |
| Headache (1/10) | |
| Insomnia (1/10) | |
| IQ < 70 (1/10) | |
| Oligophrenia (1/10) | |
| Musculoskeletal System (2/16) | Scoliosis (1/2) |
| Integumentary System (2/16) | Acne (1/2) |
| Dermatitis (1/2) | |
| Digestive System (1/16) | Dyspepsia (1/1) |
| Blood and Lymphatic Tissues (1/16) | Anemia (1/1) |
| Intermediate Metabolizers (n = 31, 44%) | |
| Nervous System (29/101) | Anxiety (2/29) |
| Dementia (4/29) | |
| Epilepsy (6/29) | |
| Insomnia (4/29) | |
| IQ < 70 (7/29) | |
| Nervous Agitation (4/29) | |
| Renal and Urinary System (12/101) | Urinary Incontinence (11/12) |
| Digestive System (11/101) | Constipation (3/11) |
| Respiratory, Thoracic, and Mediastinal System (9/101) | Bronchitis (3/9) |
| Asthma (3/9) | |
| Normal Metabolizers (n = 32, 45%) | |
| Nervous System (30/96) | Anxiety (2/30) |
| Epilepsy (4/30) | |
| Insomnia (5/30) | |
| IQ < 70 (5/30) | |
| Nervous Agitation (8/30) | |
| Integumentary System (13/96) | Dermatitis (7/13) |
| Renal and Urinary System (11/96) | Urinary Incontinence (10/11) |
| Digestive System (9/96) | Constipation (7/9) |
| Ultra-Rapid Metabolizers (n = 3; 4%) | |
| Nervous System (3/9) | Insomnia (2/3) |
| IQ < 70 (1/3) | |
| Renal and Urinary System (1/9) | Urinary Incontinence (1/1) |
| Digestive System (1/9) | Malocclusion (1/1) |
| Respiratory System (2/9) | Asthma (1/2) |
| Nasal Polyps (1/2) | |
| Vascular System (1/9) | Subclavian Artery Compression Syndrome (1/1) |
| Genetic Diseases (1/9) | Fragile X Syndrome (1/1) |
| Poor Metabolizers—Median (IQR) 4 (3–5) Drugs/Patient | ||||
|---|---|---|---|---|
| Drug | Median (IQR) | Prescribed Dosage | Main Drugs Prescribed | |
| Antipsychotic | 2 (1–3)/patient | DDR: | 100% | Quetiapine (29%, CYP3A4 substate) |
| Risperidone (21%, CYP2D6 substate) | ||||
| Levomepromazine (14%, CYP2D6 inhibitor) | ||||
| Antidepressant | 1 (0.5–1)/patient | SUPDDR: | 17% | Sertraline (33%, CYP2D6 inhibitor) |
| DDR: | 67% | Fluoxetine (17%, CYP2D6 inhibitor) | ||
| INFDDR: | 17% | Fluvoxamine (17%, CYP2D6 substate) | ||
| Anticonvulsant | 1 (0–2)/patient | DDR: | 100% | Lamotrigine (40%, NA) |
| Valproic Acid (20%, 2C19 substrate) | ||||
| Levetiracetam (20%, 2C19 substrate) | ||||
| Anxiolytic | 1 (0–2)/patient | DDR: | 100% | Lorazepam (40%, NA) |
| Lormetazepam (40%, NA) | ||||
| Clonazepam (20%, NA) | ||||
| Intermediate Metabolizers—Median (IQR) 4 (2–7) Drugs/Patient | ||||
| Drug | Median (IQR) | Prescribed Dosage | Main Drugs Prescribed | |
| Antipsychotic | 1 (1–2)/patient | SUPDDR: | 7% | Risperidone (19%, CYP2D6 substate) |
| DDR: | 93% | Levomepromazine (14%, CYP2D6 inhibitor) | ||
| INFDDR: | 0% | Olanzapine (12%, CYP1A2 substrate) | ||
| Antidepressant | 0 (0–1)/patient | DDR: | 100% | Fluvoxamine (63%, CYP2D6 substate) |
| Fluoxetine (25%, CYP2D6 inhibitor) | ||||
| Trazodone (13%, CYP3A4 substrate) | ||||
| Anticonvulsant | 1 (0–2)/patient | SUPDDR: | 15% | Topiramate (30%, CYP2C19 inhibitor) |
| DDR: | 85% | Valproic Acid (18%, CYP2C19 substrate) | ||
| INFDDR: | 0% | Carbamazepine (12%, CYP2C19 inducer) | ||
| Anxiolytic | 0 (0–1)/patient | SUPDDR: | 9% | Clorazepate (27%, NA)Clonazepam (23%, NA)Lormetazepam (18%, NA) |
| DDR: | 86% | |||
| INFDDR | 5% | |||
| Normal Metabolizers—Median (IQR) 4 (2–6) Drugs/Patient | ||||
| Drug | Median (IQR) | Prescribed Dosage | Main Drugs Prescribed | |
| Antipsychotic | 1 (1–3)/patient | SUPDDR: | 11% | Risperidone (16%, CYP2D6 substate) |
| DDR: | 88% | Levomepromazine (16%, CYP2D6 inhibitor) | ||
| INFDDR: | 1% | Olanzapine (16%, CYP1A2 substrate) | ||
| Antidepressant | 0 (0–1)/patient | SUPDDR | 0% | Fluvoxamine (35%, CYP2D6 substate) |
| DDR: | 96% | Sertraline (17%, CYP2D6 inhibitor) | ||
| INFDDR: | 4% | Trazodone (13%, CYP3A4 substrate) | ||
| Anticonvulsant | 1 (0–1)/patient | SUPDDR | 6% | Topiramate (28%, CYP2C19 inhibitor) |
| DDR: | 94% | Valproic Acid (22%, CYP2C19 substrate) | ||
| INFDDR: | 0% | Oxcarbazepine (13%, CYP3A4 inducer) | ||
| Anxiolytic | 0 (0–1)/patient | SUPDDR | 14% | Clonazepam (29%, NA) |
| DDR: | 86% | Lormetazepam (19%, NA) | ||
| INFDDR R: | 0% | Diazepam (19%, CYP2C19 inducer) | ||
| Ultra-Rapid Metabolizers—Median (IQR) 6 (3–6) Drugs/Patient | ||||
| Antipsychotic | 3 (1–4)/patient | DDR: | 100% | Haloperidol (22%, CYP2D6 substrate) |
| Olanzapine (22%, CYP1A2 substrate) | ||||
| Amisulpride/Levomepromazine/ | ||||
| Quetiapine/ | ||||
| (11%, CYP2D6 inhibitor) | ||||
| Antidepressant | 0 (0–1)/patient | DDR: | 100% | Fluoxetine (100%, CYP2D6 inhibitor) |
| Anticonvulsant | 0 (0–1)/patient | DDR: | 100% | Topiramate (100%, CYP2C19 inhibitor) |
| Anxiolytic | No reported use in UMs | - | - | |
| INFDDR | DDR | SUPDDR | p-Value | |
|---|---|---|---|---|
| Metabolizer phenotype | 0.000035 | |||
| PM | 100% | 0% | 0% | |
| EM (IM, NM) | 0% | 84.2% | 100% | |
| UM | 0% | 15.8% | 0% | |
| Drug metabolism | 0.683 | |||
| CYP2D6 substrate | 50% | 57.9% | 75% | |
| CYPD26 inhibitor | 50% | 27.3% | 0% | |
| No interaction | 0% | 15.8% | 25% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ballester, P.; Espadas, C.; Almenara, S.; Barrachina, J.; Muriel, J.; Ramos, E.; Toral, N.; Belda, C.; Peiró, A.M. CYP2D6 Genotype and Pharmacovigilance Impact on Autism Spectrum Disorder: A Naturalistic Study with Extreme Phenotype Analysis. Pharmaceuticals 2023, 16, 954. https://doi.org/10.3390/ph16070954
Ballester P, Espadas C, Almenara S, Barrachina J, Muriel J, Ramos E, Toral N, Belda C, Peiró AM. CYP2D6 Genotype and Pharmacovigilance Impact on Autism Spectrum Disorder: A Naturalistic Study with Extreme Phenotype Analysis. Pharmaceuticals. 2023; 16(7):954. https://doi.org/10.3390/ph16070954
Chicago/Turabian StyleBallester, Pura, Cristina Espadas, Susana Almenara, Jordi Barrachina, Javier Muriel, Enrique Ramos, Natalia Toral, César Belda, and Ana M. Peiró. 2023. "CYP2D6 Genotype and Pharmacovigilance Impact on Autism Spectrum Disorder: A Naturalistic Study with Extreme Phenotype Analysis" Pharmaceuticals 16, no. 7: 954. https://doi.org/10.3390/ph16070954
APA StyleBallester, P., Espadas, C., Almenara, S., Barrachina, J., Muriel, J., Ramos, E., Toral, N., Belda, C., & Peiró, A. M. (2023). CYP2D6 Genotype and Pharmacovigilance Impact on Autism Spectrum Disorder: A Naturalistic Study with Extreme Phenotype Analysis. Pharmaceuticals, 16(7), 954. https://doi.org/10.3390/ph16070954