Butremycin, the 3-Hydroxyl Derivative of Ikarugamycin and a Protonated Aromatic Tautomer of 5′-Methylthioinosine from a Ghanaian Micromonospora sp. K310
Abstract
:1. Introduction

2. Results and Discussion
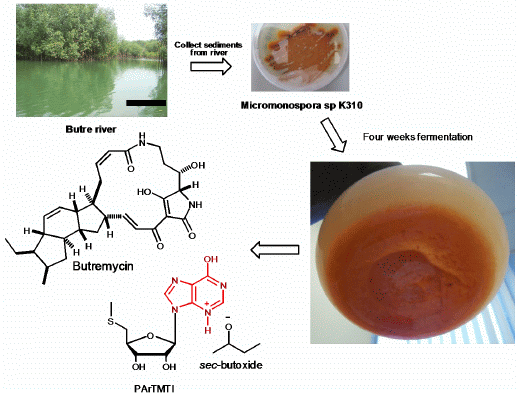
2.1. Sediment Sample Collection Sites
2.2. Taxonomy of Strain K310 (Genbank Number KF803252)

2.3. Structure Determination of Compound 2 (Butremycin)

| Position | δH mult (J Hz) | δC mult | HMBC |
|---|---|---|---|
| 1 | 195.5, C | 2 | |
| 2 | 3.70, d (2.4) | 68.4, CH | |
| 3 | 3.86, ddd (7.2, 1.9, 1.9) | 72.5, CH | 2, 4, 5 |
| 4 | 1.51, m; 1.24, m | 32.1, CH2 | 2, 3 |
| 5 | 3.42, m; 2.71, t (12.0) | 37.9, CH2 | 3 |
| 6-NH | 8.43, s | ||
| 7 | 168.9, C | 5, 8, 9 | |
| 8 | 5.76, dd (11.4, 2.8) | 124.8, CH | 10 |
| 9 | 6.02, ddd (11.4, 11.4, 2.6) | 141.8, CH | 10 |
| 10 | 3.53, m; 2.09, m | 25.9, CH2 | 8 |
| 11 | 1.35, m | 50.3, CH | 9, 10, 20, 22, 23 |
| 12 | 2.51, m | 43.4, CH | 10, 14 |
| 13 | 5.62, ddd (9.9, 2.8, 2.8) | 129.6, CH | |
| 14 | 5.83, dt (10.0, 2.1) | 131.9, CH | |
| 15 | 1.48, m | 48.4, CH | 13, 14, 19 |
| 16 | 1.29, m | 48.5, CH | 17, 30 |
| 17 | 2.20, m | 34.3, CH | 18, 29, 30 |
| 18 | 2.05, m; 0.63, ddd (11.9, 11.8, 6.8) | 39.9, CH2 | 19, 29 |
| 19 | 1.08, m | 50.4, CH | 14, 18, 21 |
| 20 | 2.00, m | 43.2, CH | 13, 18, 21 |
| 21 | 2.01, m; 1.10, m | 38.1, CH2 | 19, 22, 23 |
| 22 | 2.32, m | 49.6, CH | 21, 23, 24 |
| 23 | 6.47, dd (15.1, 10.0) | 147.8, CH | 21, 22 |
| 24 | 7.30, d (15.2) | 130.4, CH | 22 |
| 25 | 184.7, C | 23, 24 | |
| 26 | 102.8, C | ||
| 27 | 179.9, C | 2 | |
| 28-NH | 8.43, s | ||
| 29 | 0.81, d (6.0) | 18.1, CH3 | 17, 18 |
| 30 | 1.42, m; 1.30, m | 22.7, CH2 | 16, 31 |
| 31 | 0.87, t (7.1) | 13.7, CH3 | 30 |
2.4. Structure Determination of Compound 3

| Position | δH mult (J Hz) | δC mult | HMBC |
|---|---|---|---|
| 1-N | |||
| 2 | 8.07, s | 146.8, CH | |
| 3-NH | 8.29, s | ||
| 4 | 150.2, C | 2, NH-3,1′ | |
| 5 | 125.8, C | 2, NH-3 | |
| 6 | 158.9, C | 2 | |
| 7-N | |||
| 8 | 140.9, CH | 1′ | |
| 9-N | |||
| 1′ | 6.02, d (4.8) | 90.2, CH | 2′,3′ |
| 2′ | 4.73, t (5.1) | 75.2, CH | 1′ |
| 3′ | 4.32, t (4.8) | 73.9, CH | 4′,5′ |
| 4′ | 4.22, q (5.4) | 85.6, CH | 2′,5′ |
| 5′ | 2.93, dd (14.1, 5.5) 2.873, dd (14.1, 5.9) | 37.5, CH2 | 3′,7′ |
| 6′-S | |||
| 7′ | 2.14, s | 16.5, CH3 | 5′ |
2.5. Possible Biosynthesis of Compound 3

2.6. Antibacterial Activity of Butremycin
| Strain | Source of Isolate | MIC (µg mL−1) | |
|---|---|---|---|
| Butremycin | Rifampicin | ||
| S. aureus ATCC 25923 a | ATCC | 50 | 0.003 |
| E. coli ATCC 25922 a | ATCC | 50 | 0.01 |
| SMRSA 105 | Toe wound | >50 | 0.007 |
| SMRSA 124 | Open wound | >50 | 0.03 |
| SMRSA 116 | Knee abscess | >50 | 0.01–0.03 |
| EMRSA 15 | Urine infection | 50 | 0.001–0.003 |
3. Experimental Section
3.1. General Experimental Procedures
3.2. Identification of Micromonospora sp. K310
3.3. Isolation of Micromonospora sp. K310 and Preliminary Screening of the Secondary Metabolites
3.4. Large Scale Fermentation of K310
3.5. Extraction, Isolation and Purification of Compounds
3.6. Antibacterial Activity
4. Conclusions
Supplementary Files
Acknowledgments
Conflicts of Interest
References
- Hong, K.; Gao, A.-H.; Xie, Q.-Y.; Gao, H.; Zhuang, L.; Lin, H.-P.; Yu, H.-P.; Li, J.; Yao, X.-S.; Goodfellow, M.; et al. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 2009, 7, 24–44. [Google Scholar] [CrossRef]
- Maskey, R.P.; Li, F.C.; Qin, S.; Fiebig, H.H.; Laatsch, H. Chandrananimycins A–C: Production of novel anticancer antibiotics from a marine Actinomadura sp. isolate M048 by variation of medium composition and growth conditions. J. Antibiot. (Tokyo) 2003, 56, 622–629. [Google Scholar] [CrossRef]
- Rodriguez, J.C.; Fernandez Puentes, J.L.; Perez Baz, J.; Canedo, L.M. IB-00208, A new cytotoxic polycyclic xanthone produced by a marine-derived actinomadura. II. Isolation, physico-chemical properties and structure determination. J. Antibiot. (Tokyo) 2003, 56, 318–321. [Google Scholar] [CrossRef]
- Riedlinger, J.; Reicke, A.; Zahner, H.; Krismer, B.; Bull, A.T.; Maldonado, L.A.; Ward, A.C.; Goodfellow, M.; Bister, B.; Bischoff, D. Abyssomicins, inhibitors of the para-aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18–032. J. Antibiot. (Tokyo) 2004, 57, 271–279. [Google Scholar] [CrossRef]
- Stritzke, K.; Schulz, L.; Laatsch, H.; Helmke, E.; Beil, W. Novel caprolactones from a marine streptomycete. J. Nat. Prod. 2004, 67, 395–401. [Google Scholar] [CrossRef]
- Li, F.; Maskey, R.P.; Qin, S.; Sattler, I.; Fiebig, H.H.; Maier, A.; Zeeck, A.; Laatsch, H. Chinikomycins A and B: Isolation, structure elucidation, and biological activity of novel antibiotics from a marine Streptomyces sp. isolate M045. J. Nat. Prod. 2005, 68, 349–353. [Google Scholar] [CrossRef]
- Charan, R.D.; Schlingmann, G.; Janso, J.; Bernan, V.; Feng, X.; Carter, G.T. Diazepinomicin, a new antimicrobial alkaloid from marine Micromonospora sp. J. Nat. Prod. 2004, 67, 1431–1433. [Google Scholar] [CrossRef]
- Li, Y.; Huffman, J.; Li, Y.; Du, L.; Shen, Y. 3-Hydroxylation of the polycyclic tetramate macrolactam in the biosynthesis of antifungal HSAF from lysobacter enzymogenes C3. Med. Chem. Commun. 2012, 3, 982–986. [Google Scholar] [CrossRef]
- Blodgetta, J.A.V.; Ohb, D.-C.; Caoa, S.; Curried, C.R.; Kolterc, R.; Clardya, J. Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 11692–11697. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kupchan, M.S.; Britton, R.W.; Zeigler, M.F.; Sigel, C.W. Bruceantin, a new potent antileukemic simaroubolide from Brucea antidysenterica. J. Org. Chem. 1973, 38, 178–179. [Google Scholar] [CrossRef]
- Mikulaa, H.; Horkela, E.; Hansa, P.; Hametnera, C.; Fröhlicha, J. Structure and tautomerism of tenuazonic acid—A synergetic computational and spectroscopic approach. J. Hazard. Mater. 2013, 250–251, 308–317. [Google Scholar] [CrossRef]
- Parsek, M.R.; Val, D.L.; Hanzelka, B.L.; Cronan, J.E., Jr.; Greenberg, E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 1999, 96, 4360–4365. [Google Scholar]
- Cooley, M.; Chhabra, S.R.; Williams, P. N-acylhomoserine lactone-mediated quorum sensing: A twist in the tail and a blow for host immunity. Chem. Biol. 2008, 15, 1141–1147. [Google Scholar] [CrossRef]
- Guan, R.; Meng-Chiao, H.; Fröhlich, R.F.G.; Tyler, P.C.; Almo, S.C.; Schramm, V.L. Methylthioadenosine deaminase in an alternative quorum sensing pathway in Pseudomonas aeruginosa. Biochemistry 2012, 51, 9094–9103. [Google Scholar]
- Chun, J.; Goodfellow, M. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 1995, 45, 240–245. [Google Scholar] [CrossRef]
- Kim, O.-S.; Cho, Y.-J.; Lee, K.; Yoon, S.-H.; Kim, M.; Na, H.; Park, S.-C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C.R. Evolution of Protein Molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; Volume 3, pp. 21–132. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogeny: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Domenech, P.; Kobayashi, H.; LeVier, K.; Walker, G.C.; Barry, C.E. BacA, an ABC transporter involved in maintenance of chronic murine infections with Mycobacterium tuberculosis. J. Bacteriol. 2009, 191, 477–485. [Google Scholar] [CrossRef]
- Müller, H.J.; Hinton, J. A protein-free medium for primary isolation of the Gonococcus and Meningococcus. Proc. Soc. Exp. Biol. Med. 1941, 48, 330–333. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kyeremeh, K.; Acquah, K.S.; Sazak, A.; Houssen, W.; Tabudravu, J.; Deng, H.; Jaspars, M. Butremycin, the 3-Hydroxyl Derivative of Ikarugamycin and a Protonated Aromatic Tautomer of 5′-Methylthioinosine from a Ghanaian Micromonospora sp. K310. Mar. Drugs 2014, 12, 999-1012. https://doi.org/10.3390/md12020999
Kyeremeh K, Acquah KS, Sazak A, Houssen W, Tabudravu J, Deng H, Jaspars M. Butremycin, the 3-Hydroxyl Derivative of Ikarugamycin and a Protonated Aromatic Tautomer of 5′-Methylthioinosine from a Ghanaian Micromonospora sp. K310. Marine Drugs. 2014; 12(2):999-1012. https://doi.org/10.3390/md12020999
Chicago/Turabian StyleKyeremeh, Kwaku, Kojo Sekyi Acquah, Anil Sazak, Wael Houssen, Jioji Tabudravu, Hai Deng, and Marcel Jaspars. 2014. "Butremycin, the 3-Hydroxyl Derivative of Ikarugamycin and a Protonated Aromatic Tautomer of 5′-Methylthioinosine from a Ghanaian Micromonospora sp. K310" Marine Drugs 12, no. 2: 999-1012. https://doi.org/10.3390/md12020999
APA StyleKyeremeh, K., Acquah, K. S., Sazak, A., Houssen, W., Tabudravu, J., Deng, H., & Jaspars, M. (2014). Butremycin, the 3-Hydroxyl Derivative of Ikarugamycin and a Protonated Aromatic Tautomer of 5′-Methylthioinosine from a Ghanaian Micromonospora sp. K310. Marine Drugs, 12(2), 999-1012. https://doi.org/10.3390/md12020999