Chemoinformatic Analysis as a Tool for Prioritization of Trypanocidal Marine Derived Lead Compounds
Abstract
:1. Introduction
2. Results and Discussion

2.1. Marine Fraction Library

2.2. Taxonomic Origin


2.3. Isolation of Marine Natural Products

2.4. Antitrypanosomal Activities
2.5. Lipinski’s Rule-of-Five
| Compound | MW | Log D5.5 | Log P | HBA | HBD | %PSA | No. of Violations |
|---|---|---|---|---|---|---|---|
| 1 | 368.51 | 4.37 | 5.29 | 5 | 1 | 11.28 | 1 |
| 2 | 344.44 | 2.40 | 4.33 | 6 | 2 | 15.65 | 0 |
| 3 | 352.47 | 4.20 | 4.20 | 4 | 0 | 10.28 | 0 |
| 4 | 406.56 | 4.12 | 5.36 | 5 | 1 | 10.54 | 1 |
| 5 | 406.56 | 3.30 | 4.82 | 5 | 1 | 9.58 | 0 |
| 6 | 416.55 | 5.60 | 5.60 | 4 | 0 | 7.76 | 1 |
| 7 | 406.56 | 3.37 | 4.98 | 5 | 1 | 9.60 | 0 |
| 8 | 473.04 | −1.64 | 4.05 | 3 | 1 | 4.88 | 0 |
| 9 | 470.02 | 0.29 | 0.29 | 2 | 0 | 3.37 | 0 |
| 10 | 341.36 | 0.76 | 2.55 | 5 | 3 | 20.20 | 0 |
| 11 | 443.29 | 1.61 | 3.72 | 4 | 3 | 17.46 | 0 |
| 12 | 283.28 | 2.98 | 2.98 | 4 | 0 | 16.07 | 0 |
| 13 | 356.38 | 4.70 | 4.70 | 4 | 0 | 11.78 | 0 |
| 14 | 269.30 | 3.15 | 3.40 | 3 | 1 | 10.92 | 0 |
| 15 | 495.17 | 0.66 | 1.31 | 8 | 6 | 28.51 | 1 |
| 16 | 622.15 | 2.85 | 3.57 | 6 | 3 | 16.26 | 1 |
| 17 | 747.07 | 0.65 | 3.74 | 8 | 3 | 15.77 | 1 |
| 18 | 898.19 | −0.09 | 0.62 | 11 | 4 | 21.16 | 2 |
| 19 | 334.39 | −1.43 | −1.44 | 3 | 2 | 12.63 | 0 |
| 20 | 321.37 | −0.94 | 1.82 | 4 | 2 | 14.92 | 0 |
| 21 | 332.38 | −0.75 | −0.75 | 2 | 2 | 13.52 | 0 |
| 22 | 320.37 | −1.65 | −1.67 | 3 | 3 | 16.28 | 0 |
| 23 | 322.38 | −1.76 | −1.76 | 3 | 3 | 15.17 | 0 |
| 24 | 254.29 | 1.39 | 1.39 | 3 | 2 | 18.44 | 0 |
| 25 | 255.27 | 1.22 | 1.22 | 2 | 1 | 16.58 | 0 |
| 26 | 268.31 | 1.61 | 1.61 | 3 | 1 | 13.69 | 0 |
| 27 | 333.18 | 2.15 | 2.15 | 3 | 2 | 17.44 | 0 |
| 28 | 324.13 | −0.83 | −0.83 | 4 | 5 | 35.30 | 0 |
| 29 | 245.24 | −1.30 | −1.30 | 4 | 5 | 37.79 | 0 |
| 30 | 389.05 | −0.80 | 1.55 | 4 | 2 | 21.89 | 0 |
| 31 | 421.66 | 3.69 | 6.89 | 3 | 2 | 7.56 | 1 |
| 32 | 420.65 | 3.33 | 6.53 | 2 | 2 | 8.43 | 1 |
| 33 | 420.65 | 3.33 | 6.53 | 2 | 2 | 8.43 | 1 |
| 34 | 470.73 | 1.67 | 6.81 | 4 | 0 | 4.87 | 1 |
| 35 | 472.75 | 2.16 | 7.15 | 4 | 0 | 3.86 | 1 |
| 36 | 381.64 | −2.06 | 4.81 | 3 | 1 | 4.60 | 0 |
| 37 | 227.39 | 0.73 | 3.74 | 2 | 2 | 10.30 | 0 |
| 38 | 422.63 | 1.47 | 1.47 | 3 | 1 | 8.50 | 0 |
| 39 | 443.45 | 4.81 | 4.82 | 5 | 3 | 18.18 | 0 |
| 40 | 285.34 | 2.93 | 3.83 | 1 | 2 | 11.36 | 0 |

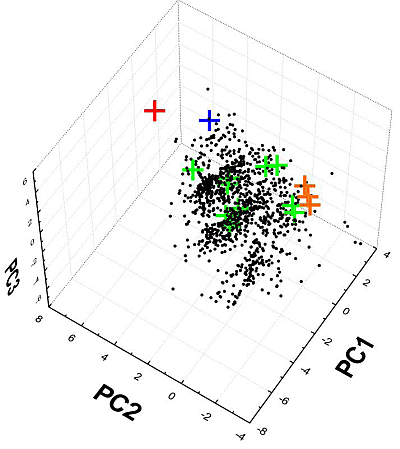
2.6. Chemical Clustering

2.7. ChemGPS-NP Analysis

3. Experimental Section
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Construction of Fraction Library
3.4. Extraction and Isolation
3.5. Cheminformatics
4. Conclusions
Acknowledgements
Author Contributions
Conflicts of Interest
References
- WHO (World Health Organisation) Trypanosomiasis, Human African (sleeping sickness). Available online: http://www.who.int/mediacentre/factsheets/fs259/en/ (accessed on 20 June 2013).
- Bouteille, B.; Oukem, O.; Bisser, S.; Dumas, M. Treatment perspectives for human African trypanosomiasis. Fundam. Clin. Pharmacol. 2003, 17, 171–181. [Google Scholar] [CrossRef]
- Matovu, E.; Stewart, M.L.; Geiser, F.; Brun, R.; Maser, P.; Wallace, L.J.; Burchmore, R.J.; Enyaru, J.C.; Barrett, M.P.; Kaminsky, R.; et al. Mechanisms of arsenical and diamidine uptake and resistance in Trypanosoma brucei. Eukaryot. Cell 2003, 2, 1003–1008. [Google Scholar] [CrossRef]
- Mallari, J.P.; Shelat, A.A.; Obrien, T.; Caffrey, C.R.; Kosinski, A.; Connelly, M.; Harbut, M.; Greenbaum, D.; McKerrow, J.H.; Guy, R.K. Development of potent purine-derived nitrile inhibitors of the trypanosomal protease TbcatB. J. Med. Chem. 2008, 51, 545–552. [Google Scholar] [CrossRef]
- DNDi (Drugs for Neglected Diseases initiative), Fexinidazole. Available online: http://www.dndi.org/diseases-projects/portfolio/fexinidazole.html (accessed on 20 June 2013).
- Salem, M.M.; Werbovetz, K.A. Natural products from plants as drug candidates and lead compounds against leishmaniasis and trypanosomiasis. Curr. Med. Chem. 2006, 13, 2571–2598. [Google Scholar] [CrossRef]
- Kossuga, M.H.; Nascimento, A.M.; Reimao, J.Q.; Tempone, A.G.; Taniwaki, N.N.; Veloso, K.; Ferreira, A.G.; Cavalcanti, B.C.; Pessoa, C.; Moraes, M.O.; et al. Antiparasitic, antineuroinflammatory, and cytotoxic polyketides from the marine sponge Plakortis angulospiculatus collected in Brazil. J. Nat. Prod. 2008, 71, 334–339. [Google Scholar] [CrossRef]
- Rubio, B.K.; Tenney, K.; Ang, K.H.; Abdulla, M.; Arkin, M.; McKerrow, J.H.; Crews, P. The marine sponge Diacarnus bismarckensis as a source of peroxiterpene inhibitors of Trypanosoma brucei, the causative agent of sleeping sickness. J. Nat. Prod. 2009, 72, 218–222. [Google Scholar] [CrossRef]
- Feng, Y.; Davis, R.A.; Sykes, M.L.; Avery, V.M.; Quinn, R.J. Iotrochamides A and B, antitrypanosomal compounds from the Australian marine sponge Iotrochota sp. Bioorg. Med. Chem. Lett. 2012, 22, 4873–4876. [Google Scholar] [CrossRef]
- Feng, Y.J.; Davis, R.A.; Sykes, M.L.; Avery, V.M.; Carroll, A.R.; Camp, D.; Quinn, R.J. Antitrypanosomal pyridoacridine alkaloids from the Australian ascidian Polysyncraton echinatum. Tetrahedron Lett. 2010, 51, 2477–2479. [Google Scholar] [CrossRef]
- Feng, Y.; Davis, R.A.; Sykes, M.; Avery, V.M.; Camp, D.; Quinn, R.J. Antitrypanosomal cyclic polyketide peroxides from the Australian marine sponge Plakortis sp. J. Nat. Prod. 2010, 73, 716–719. [Google Scholar] [CrossRef]
- Feng, Y.J.; Davis, R.A.; Sykes, M.L.; Avery, V.M.; Camp, D.; Quinn, R.J. Pseudoceratinazole A: A novel bromotyrosine alkaloid from the Australian sponge Pseudoceratina sp. Tetrahedron Lett. 2010, 51, 4847–4850. [Google Scholar] [CrossRef]
- Davis, R.A.; Sykes, M.; Avery, V.M.; Camp, D.; Quinn, R.J. Convolutamines I and J, antitrypanosomal alkaloids from the bryozoan Amathia tortusa. Bioorg. Med. Chem. 2011, 19, 6615–6619. [Google Scholar] [CrossRef]
- Camp, D.; Davis, R.A.; Campitelli, M.; Ebdon, J.; Quinn, R.J. Drug-like properties: Guiding principles for the design of natural product libraries. J. Nat. Prod. 2012, 75, 72–81. [Google Scholar] [CrossRef]
- Camp, D.; Campitelli, M.; Carroll, A.R.; Davis, R.A.; Quinn, R.J. Front-loading natural-product-screening libraries for log P: Background, development, and implementation. Chem. Biodivers. 2013, 10, 524–537. [Google Scholar] [CrossRef]
- Ibrahim, S.R.; Ebel, R.; Wray, V.; Muller, W.E.; Edrada-Ebel, R.; Proksch, P. Diacarperoxides, norterpene cyclic peroxides from the sponge Diacarnus megaspinorhabdosa. J. Nat. Prod. 2008, 71, 1358–1364. [Google Scholar] [CrossRef]
- Capon, R.J.; Macleod, J.K.; Willis, A.C. Trunculin-A and trunculin-B, norsesterterpene cyclic peroxides from a Marine Sponge, Latrunculia brevis. J. Org. Chem. 1987, 52, 339–342. [Google Scholar] [CrossRef]
- Ovenden, S.P.B.; Capon, R.J. Trunculins G–I: New norsesterterpene cyclic peroxides from a southern Australian marine sponge, Latrunculia sp. Aust. J. Chem. 1998, 51, 573–579. [Google Scholar] [CrossRef]
- Butler, M.S.; Capon, R.J. Trunculin-F and contrunculin-A and contrunculin-B—novel oxygenated norterpenes from a southern Australian marine sponge, Latrunculia conulosa. Aust. J. Chem. 1993, 46, 1363–1374. [Google Scholar] [CrossRef]
- Kobayashi, J.; Honma, K.; Sasaki, T.; Tsuda, M. Purealidins J–R, new bromotyrosine alkaloids from the Okinawan marine sponge Psammaplysilla purea. Chem. Pharm. Bull. 1995, 43, 403–407. [Google Scholar] [CrossRef]
- Yagi, H.; Matsunaga, S.; Fusetani, N. Bioactive marine metabolites. 49. Purpuramines A–I, new bromotyrosine-derived metabolites from the marine sponge Psammaplysilla purpurea. Tetrahedron 1993, 49, 3749–3754. [Google Scholar] [CrossRef]
- Davis, R.A.; Buchanan, M.S.; Duffy, S.; Avery, V.M.; Charman, S.A.; Charman, W.N.; White, K.L.; Shackleford, D.M.; Edstein, M.D.; Andrews, K.T.; et al. Antimalarial activity of pyrroloiminoquinones from the Australian marine sponge Zyzzya sp. J. Med. Chem. 2012, 55, 5851–5858. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Harper, M.K.; Faulkner, D.J. Makaluvamines H–M and damirone C from the Pohnpeian sponge Zyzzya fuliginosa. J. Nat. Prod. 1995, 58, 1861–1867. [Google Scholar] [CrossRef]
- Carney, J.R.; Scheuer, P.J.; Kelly-Borges, M. Makaluvamine G, a cytotoxic pigment from an Indonesian sponge Histodermella sp. Tetrahedron 1993, 49, 8483–8486. [Google Scholar]
- Kochanowska, A.J.; Rao, K.V.; Childress, S.; El-Alfy, A.; Matsumoto, R.R.; Kelly, M.; Stewart, G.S.; Sufka, K.J.; Hamann, M.T. Secondary metabolites from three Florida sponges with antidepressant activity. J. Nat. Prod. 2008, 71, 186–189. [Google Scholar] [CrossRef]
- Hu, J.F.; Schetz, J.A.; Kelly, M.; Peng, J.N.; Ang, K.K.H.; Flotow, H.; Leong, C.Y.; Ng, S.B.; Buss, A.D.; Wilkins, S.P.; Hamann, M.T. New antiinfective and human 5-HT2 receptor binding natural and semisynthetic compounds from the Jamaican sponge Smenospongia aurea. J. Nat. Prod. 1977, 40, 479–481. [Google Scholar]
- Annoura, H.; Tatsuoka, T. Total syntheses of hymenialdisine and debromohymenialdisine: Stereospecific construction of the 2-amino-4-oxo-2-imidazolin-5(Z)-disubstituted ylidene ring system. Tetrahedron Lett. 1995, 36, 413–416. [Google Scholar] [CrossRef]
- Sharma, G.; Magdoff-Fairchild, B. Natural products of marine sponges. 7. The constitution of weakly basic guanidine compounds, dibromophakellin and monobromophakellin. J. Org. Chem. 1977, 42, 4118–4124. [Google Scholar] [CrossRef]
- Davis, R.A.; Carroll, A.R.; Quinn, R.J. Lepadins F–H, new cis-decahydroquinoline alkaloids from the Australian ascidian Aplidium tabascum. J. Nat. Prod. 2002, 65, 454–457. [Google Scholar] [CrossRef]
- Wright, A.D.; Goclik, E.; Koenig, G.M.; Kaminsky, R. Lepadins D–F: Antiplasmodial and antitrypanosomal decahydroquinoline derivatives from the tropical marine tunicate Didemnum sp. J. Med. Chem. 2002, 45, 3067–3072. [Google Scholar] [CrossRef]
- Goud, T.V.; Reddy, N.S.; Swamy, N.R.; Ram, T.S.; Venkateswarlu, Y. Anti-HIV active petrosins from the marine sponge Petrosia similis. Bio. Pharm. Bull. 2003, 26, 1498–1501. [Google Scholar] [CrossRef]
- Kobayashi, M.; Kawazoe, K.; Kitagawa, I. Chemical structures and optical properties of dimeric quinolizidine-alkaloid macrocycles isolated from an marine sponge. Tennen Yuki Kagobutsu Toronkai Koen Yoshishu 1989, 31, 332–339. [Google Scholar]
- Braekman, J.C.; Daloze, D.; Macedo de Abreu, P.; Piccinni-Leopardi, C.; Germain, G.; Van Meerssche, M. A novel type of bisquinolizidine alkaloid from the sponge Petrosia seriata. Tetrahedron Lett. 1982, 23, 4277–4280. [Google Scholar] [CrossRef]
- Clark, R.J.; Garson, M.J.; Hooper, J.N.A. Antifungal alkyl amino alcohols from the tropical marine sponge Haliclona sp. J. Nat. Prod. 2001, 64, 1568–1571. [Google Scholar] [CrossRef]
- Schmitz, F.J.; Hollenbeak, K.H.; Prasad, R.S. Marine natural-products—cytotoxic spermidine derivatives from the soft coral Sinularia brongersmai. Tetrahedron Lett. 1979, 20, 3387–3390. [Google Scholar]
- Hertiani, T.; Edrada-Ebel, R.; Ortlepp, S.; van Soest, R.W.M.; de Voogd, N.J.; Wray, V.; Hentschel, U.; Kozytska, S.; Mueller, W.E.G.; Proksch, P. From anti-fouling to biofilm inhibition: New cytotoxic secondary metabolites from two Indonesian Agelas sponges. Bioorg. Med. Chem. 2010, 18, 1297–1311. [Google Scholar] [CrossRef]
- Zhang, H.; Conte, M.M.; Huang, X.C.; Khalil, Z.; Capon, R.J. A search for BACE inhibitors reveals new biosynthetically related pyrrolidones, furanones and pyrroles from a southern Australian marine sponge, Ianthella sp. Org. Biomol. Chem. 2012, 10, 2656–2663. [Google Scholar] [CrossRef]
- Kang, H.; Fenical, W. New isoeudistomin class dihydro-β-carbolines from an undescribed ascidian of the genus Eudistoma. Nat. Prod. Lett. 1996, 9, 7–12. [Google Scholar] [CrossRef]
- Holla, H.; Labaied, M.; Pham, N.; Jenkins, I.D.; Stuart, K.; Quinn, R.J. Synthesis of antitrypanosomal 1,2-dioxane derivatives based on a natural product scaffold. Bioorg. Med. Chem. Lett. 2011, 21, 4793–4797. [Google Scholar] [CrossRef]
- Instant JChem, version 6.03; ChemAxon Kft: Budapest, Hungary, 2013.
- Schrödinger Release 2013-1: Canvas, version 1.6; Schrödinger, LLC: New York, NY, USA, 2013.
- Rosén, J.; Lövgren, A.; Kogej, T.; Muresan, S.; Gottfries, J.; Backlund, A. ChemGPS-NP(Web): Chemical space navigation online. J. Comput. Aided Mol. Des. 2009, 23, 253–259. [Google Scholar] [CrossRef]
- Larsson, J.; Gottfries, J.; Muresan, S.; Backlund, A. ChemGPS-NP: Tuned for navigation in biologically relevant chemical space. J. Nat. Prod. 2007, 70, 789–794. [Google Scholar] [CrossRef]
- Knox, C.; Law, V.; Jewison, T.; Liu, P.; Ly, S.; Frolkis, A.; Pon, A.; Banco, K.; Mak, C.; Neveu, V.; et al. DrugBank 3.0: A comprehensive resource for “omics” research on drugs. Nucleic Acids Res. 2011, 39, D1035–D1041. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008, 36, D901–D906. [Google Scholar]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Feng, Y.; Campitelli, M.; Davis, R.A.; Quinn, R.J. Chemoinformatic Analysis as a Tool for Prioritization of Trypanocidal Marine Derived Lead Compounds. Mar. Drugs 2014, 12, 1169-1184. https://doi.org/10.3390/md12031169
Feng Y, Campitelli M, Davis RA, Quinn RJ. Chemoinformatic Analysis as a Tool for Prioritization of Trypanocidal Marine Derived Lead Compounds. Marine Drugs. 2014; 12(3):1169-1184. https://doi.org/10.3390/md12031169
Chicago/Turabian StyleFeng, Yunjiang, Marc Campitelli, Rohan A. Davis, and Ronald J. Quinn. 2014. "Chemoinformatic Analysis as a Tool for Prioritization of Trypanocidal Marine Derived Lead Compounds" Marine Drugs 12, no. 3: 1169-1184. https://doi.org/10.3390/md12031169
APA StyleFeng, Y., Campitelli, M., Davis, R. A., & Quinn, R. J. (2014). Chemoinformatic Analysis as a Tool for Prioritization of Trypanocidal Marine Derived Lead Compounds. Marine Drugs, 12(3), 1169-1184. https://doi.org/10.3390/md12031169