Sterols from the Green Alga Ulva australis
Abstract
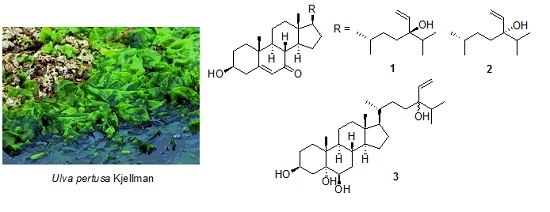
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Algal Material
3.3. Extraction and Isolation
3.4. Human Recombinant Aldose Reductase Inhibitory Activity Assay
4. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 33, 382–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed]
- Sakai, R.; Swanson, G.T. Recent progress in neuroactive marine natural products. Nat. Prod. Rep. 2014, 31, 273–309. [Google Scholar] [CrossRef] [PubMed]
- Sunassee, S.N.; Davies-Coleman, M.T. Cytotoxic and antioxidant marine prenylated quinones and hydroquinones. Nat. Prod. Rep. 2012, 29, 513–535. [Google Scholar] [CrossRef] [PubMed]
- Lahaye, M.; Jegou, D. Chemical and physical-chemical characteristics of dietary fibers from Ulva lactuca (L.) Thuret and Enteromorpha compressa (L.) Grev. J. Appl. Phycol. 1993, 5, 195–200. [Google Scholar] [CrossRef]
- Ali, I.; Manzoor, Z.; Koo, J.E.; Kim, J.E.; Byeon, S.H.; Yoo, E.S.; Kang, H.K.; Hyun, J.W.; Lee, N.H.; Koh, Y.S. 3-Hydroxy-4,7-megastigmadien-9-one, isolated from Ulva pertusa, attenuates TLR9-mediated inflammatory response by down-regulating mitogen-activated protein kinase and NF-κB pathways. Pharm. Biol. 2016, 55, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.Z.; Li, N.; Liu, X.G.; Zhou, G.F.; Zhang, Q.B.; Li, P.C. Antihyperlipidemic effects of different molecular weight sulfated polysaccharides from Ulva pertusa (Chlorophyta). Pharmacol. Res. 2003, 48, 543–549. [Google Scholar]
- Zeng, C.; Tseng, C.K.; Zhang, J.; Chang, C.F. Chinese seaweeds in herbal medicine. Hydrobiology 1984, 116–117, 152–155. [Google Scholar]
- Qi, H.M.; Huang, L.Y.; Liu, X.L.; Liu, D.M.; Zhang, Q.B.; Liu, S.M. Antihyperlipidemic activity of high sulfate content derivative of polysaccharide extracted from Ulva pertusa (Chlorophyta). Carbohydr. Polym. 2012, 87, 1637–1640. [Google Scholar] [CrossRef]
- Qi, H.M.; Zhang, Q.B.; Zhao, T.T.; Chen, R.; Zhang, H.; Niu, X.Z.; Li, Z.E. Antioxidant activity of different sulfate content derivatives of polysaccharide extracted from Ulva pertusa (Chlorophyta) in vitro. Int. J. Biol. Macromol. 2005, 37, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Chen, X.L.; Liu, X.D.; Zhang, F.B.; Hu, L.F.; Yue, Y.; Li, K.C.; Li, P.C. Characterization and comparison of the structural features, immune-modulatory and anti-avian influenza virus activities conferred by three algal sulfated polysaccharides. Mar. Drugs 2016, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Chu, Z.Y.; Bao, L.L.; Cheng, X.; Liang, X.F.; Liu, X.J.; Zhou, X. Effect of polysaccharide from Ulva pertusa on immune function in mice. Chin. J. Biochem. Pharm. 2006, 27, 276–279. [Google Scholar]
- Shi, J.M.; Cheng, C.L.; Zhao, H.T.; Jing, J.; Gong, N.; Lu, W.H. In vivo anti-radiation activities of the Ulva pertusa polysaccharides and polysaccharide-iron (III) complex. Int. J. Biol. Macromol. 2013, 60, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.H.; Lu, Q.H.; Zhao, Y. Studies on the chemical constituents of green algae Ulva pertusa. China J. Chin. Mater. Med. 2007, 32, 1536–1538. [Google Scholar]
- Lu, Q.H.; Xu, J.H.; Zhao, Y.; Zhou, C.X. Studies on chemical constituents of Ulva pertusa. Chin. Pharm. J. 2008, 43, 582–584. [Google Scholar]
- Wang, C.Y.; Han, L.; Kang, K.; Shao, C.L.; Wei, Y.X.; Zheng, C.J.; Guan, H.S. Secondary metabolites from green algae Ulva pertusa. Chem. Nat. Comp. 2010, 46, 828–830. [Google Scholar] [CrossRef]
- Ali, I.; Manzoor, Z.; Koh, Y.S. 3-Hydroxy-4,7-megastigmadien-9-one, isolated from Ulva pertusa, inhibits LPS-induces inflammatory response by down-regulating mitogen-activated protein kinase and NF-κB pathways. J. Bacteriol. Virol. 2016, 46, 167–172. [Google Scholar] [CrossRef]
- Wang, W.; Okada, Y.; Shi, H.B.; Wang, Y.Q.; Okuyama, T. Structures and aldose reductase inhibitory effects of bromophenols from the red alga Symphyocladia latiuscula. J. Nat. Prod. 2005, 68, 620–622. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.N.; Choi, S.H.; Moon, H.E.; Park, J.J.; Jung, H.A.; Woo, M.H.; Woo, H.C.; Choi, J.S. The inhibitory activities of the edible green alga Capsosiphon fulvescens on rat lens aldose reductase and advanced glycation end products formation. Eur. J. Nutr. 2014, 55, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Catalan, C.A.; Kokke, W.C.M.C.; Duque, C.; Djerassi, C. Synthesis of (24R)- and (24S)-5,28-stigmastadien-3β-ol and determination of the stereochemistry of their 24-hydroxy analogues, the saringosterols. J. Org. Chem. 1983, 48, 5207–5214. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, J.; Fu, Z.F.; Ye, C.; Zhang, R.S.; Song, Y.Y.; Zhang, Y.; Li, H.H.; Ying, H.; Liu, H.B. 24(S)-Saringosterol from edible marine seaweed Sargassum fusiforme is a novel selective LXRβ agonist. J. Agric. Food Chem. 2014, 62, 6130–6137. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, V.U.; Aliya, R.; Perveen, S.; Shameel, M. Sterols from marine alga Codium decorticatum. Phytochemistry 1993, 33, 1189–1192. [Google Scholar] [CrossRef]
- Wang, W.; Li, H.Y.; Wang, Y.Y.; Xia, X.; Okada, Y.; Okuyama, T. Chemical constituents from brown alga Sargassum fusiforme. Chin. Tradit. Herb. Drugs 2008, 39, 657–661. [Google Scholar]
- Das, B.; Srinivas, K.V.N.S. Studies on marine chemicals, part IV. Isolation of cholesterol derivatives from the marine sponge Spirastrella inconstans. J. Nat. Prod. 1992, 55, 1310–1312. [Google Scholar] [CrossRef]
- Notano, G.; Piccialli, V.; Sica, D. 3β,5α,6β-Trihydroxylated sterols with a saturated nucleus from two populations of the marine sponge Cliona copiosa. J. Nat. Prod. 1991, 54, 1570–1575. [Google Scholar] [CrossRef]
- Das, B.; Rao, S.P.; Srinivas, K.V.N.S. Studies on marine chemicals, part VI. A new clionasterol derivative from the marine sponge Spirastrella inconstans. J. Nat. Prod. 1992, 56, 2210–2211. [Google Scholar] [CrossRef]
- Nicotra, F.; Toma, L. A convenient procedure for the conversion of fucosterol into isofucosterol. Gazz. Chim. Itali. 1980, 110, 579–580. [Google Scholar]
- Tang, H.F.; Yi, Y.H.; Yao, X.S.; Zhou, D.Z.; Lv, T.S.; Jiang, Y.P. Studies on the bioactive steriod constituents from Sargassum carpophyllum. Chin. Pharm. J. 2002, 37, 262–265. [Google Scholar]
- Grewal, A.S.; Bhardwaj, S.; Pandita, D.; Lather, V.; Sekhon, B.S. Updates on aldose reductase inhibitors for management of diabetic complications and non-diabetic diseases. Mini-Rev. Med. Chem. 2016, 16, 120–162. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Ryu, H.S.; Rodriguez, J.P.; Lee, S. Aldose reductase inhibitory activity of quercetin from the stem of Rhododendron mucronulatum for. albiflorum. J. Appl. Biol. Chem. 2017, 60, 29–33. [Google Scholar] [CrossRef]
- Kim, S.B.; Hwang, S.H.; Suh, H.W.; Lim, S.S. Phytochemical analysis of Agrimonia pilosa Ledeb, its antioxidant activity and aldose reductase inhibitory potential. Int. J. Mol. Sci. 2017, 18, 379. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Hao, M.; Li, C.C.; Wu, W.Y.; Wang, W.W.; Ma, Z.X.; Yang, T.T.; Zhang, N.; Isaac, A.T.; Zhu, X.; et al. Quercetin inhibited epithelial mesenchymal transition in diabetic rats, high-glucose-cultured lens, and SRA01/04 cells through transforming growth factor-β2/phosphoinositide 3-kinase/Akt pathway. Mol. Cell. Endocrinol. 2017, 452, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, V.U.; Ali, M.S. Terpenoids from marine red alga Laurencia pinnatifida. Phytochemistry 1991, 30, 4172–4174. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, X.; Wen, J.W.; Chen, M.S.; He, K.J.; Liu, B.M. Chemical constituents of Pilea sinofasciata. Chin. Tradit. Herb. Drugs 2016, 47, 3159–3163. [Google Scholar]
- Sakamoto, B.; Hokama, Y.; Horgen, F.D.; Scheuer, P.J.; Kan, Y.; Nagai, H. Isolation of a sulfoquinovosyl monoacylglycerol from Bryopsis sp. (Chlorophyta): Identification of a factor causing a possible species-specific ecdysis response in Gambierdiscus toxicus (Dinophyceae). J. Phycol. 2000, 36, 924–931. [Google Scholar] [CrossRef]
- Murakami, N.; Morimito, T.; Imamura, H.; Nagatsu, A.; Sakakibara, J. Enzymatic transformation of glyceroglycolipids into sn-1 and sn-2 lysoglyceroglycolipids by use of Rhizopus arrhizus lipase. Tetrahedron 1994, 50, 1993–2002. [Google Scholar] [CrossRef]
- Doi, J.T.; Goodrow, M.H.; Musker, W.K. Neighboring group participation in organic redox reactions. 11. Anchimeric assistance by the carboxylate anion in aqueous iodine oxidations of 3-(alkylthio)propanoates. J. Org. Chem. 1986, 51, 1026–1029. [Google Scholar] [CrossRef]
- Wang, X.F.; Li, C.; Zheng, Y.Y.; Di, D.L. Polyphenols from leaves of Olea europaea. Chin. Tradit. Herb. Drugs 2011, 42, 848–851. [Google Scholar]
- Wang, X.J.; Xie, X.; Luo, X.; Song, Y.L.; Zhao, Y.W.; Huang, W.Z.; Wang, Z.Z.; Xiao, W. Chemical constituents from Rhodiola wallichiana var. cholaensis (I). Chin. Tradit. Herb. Drugs 2015, 46, 3471–3474. [Google Scholar]
- Kimura, J.; Maki, N. New loliolide derivative from the brown alga Undaria ponnatifida. J. Nat. Prod. 2002, 65, 57–58. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.R.; Ma, Q.Y.; Huang, S.Z.; Yu, Z.F.; Zhao, Y.X. Chemical constituents from stems of Cipadessa cinerascens. Chin. Tradit. Herb. Drugs 2015, 46, 633–638. [Google Scholar]
- Wang, Q.; Xu, Y.Y. Chemical constituents of Aganosma marginata. Chin. Tradit. Herb. Drugs 2015, 46, 1742–1748. [Google Scholar]
- Lin, J.B.; Zhao, L.C.; Guo, J.Z.; Liu, L.; Kuang, Y.; Yang, S.X. Chemical constituents from aerial parts of Fagopyrum dibotrys. Chin. Tradit. Herb. Drugs 2016, 47, 1841–1844. [Google Scholar]
- Suzuki, Y.; Seki, T.; Tanaka, S.; Kitamura, M. Intramolecular tsuji-trost-type allylation of carboxylic acids: Asymmetric synthesis of highly π-allyl donative lactones. J. Am. Chem. Soc. 2015, 137, 9539–9542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Xu, X.Z.; Li, C. Fructosides from Cynomorium songaricum. Phytochemistry 1996, 41, 975–976. [Google Scholar] [CrossRef]
- Tang, Y.P.; Yu, B.; Hu, J.; Wu, T.; Hui, Y.Z. The chemical constituents from the bulbs of Ornithogalum caudatum. J. Chin. Pharm. Sci. 2001, 10, 169–171. [Google Scholar]



| Position | 1 | 2 | 3 | a 1 | b 2 | c 3 | d 4 |
|---|---|---|---|---|---|---|---|
| 1 | 36.3 | 36.3 | 30.8 | 36.3 | 37.3 | 37.3 | 30.2 |
| 2 | 31.1 | 31.2 | 32.3 | 31.1 | 32.0 | 32.0 | 33.3 |
| 3 | 70.4 | 70.5 | 67.5 | 70.4 | 71.8 | 71.8 | 66.8 |
| 4 | 41.7 | 41.8 | 40.7 | 41.7 | 42.4 | 42.4 | 39.7 |
| 5 | 164.6 | 164.8 | 75.9 | 164.9 | 140.7 | 140.7 | 75.0 |
| 6 | 125.8 | 126.0 | 75.9 | 126.1 | 121.6 | 121.6 | 75.4 |
| 7 | 201.6 | 201.9 | 34.5 | 202.5 | 31.7 | 31.7 | 35.4 |
| 8 | 45.3 | 45.4 | 30.1 | 45.3 | 32.0 | 32.0 | 30.0 |
| 9 | 49.8 | 49.9 | 45.8 | 49.9 | 50.2 | 50.2 | 45.0 |
| 10 | 38.2 | 38.2 | 38.2 | 38.6 | 36.6 | 36.6 | 37.7 |
| 11 | 21.2 | 21.2 | 21.2 | 21.1 | 21.2 | 21.1 | 20.8 |
| 12 | 38.6 | 38.6 | 39.8 | 38.6 | 39.8 | 39.8 | 39.1 |
| 13 | 43.0 | 43.1 | 42.7 | 43.0 | 42.4 | 42.3 | 42.3 |
| 14 | 49.8 | 49.9 | 55.8 | 49.8 | 56.8 | 56.8 | 55.9 |
| 15 | 26.2 | 26.2 | 24.0 | 26.2 | 24.3 | 24.3 | 22.2 |
| 16 | 29.6 | 29.7 | 28.14/28.09 | 29.3 | 28.2 | 28.3 | 27.6 |
| 17 | 54.4 | 54.5 | 55.8 | 54.6 | 55.9 | 55.9 | 55.6 |
| 18 | 11.9 | 12.0 | 12.1 | 11.6 | 11.9 | 11.9 | 11.6 |
| 19 | 17.3 | 17.3 | 16.8 | 17.2 | 19.5 | 19.4 | 16.1 |
| 20 | 36.0 | 36.0 | 36.0 | 35.4 | 36.0 | 36.0 | 35.8 |
| 21 | 18.9 | 18.9 | 18.7 | 18.7 | 18.9 | 18.8 | 18.2 |
| 22 | 29.0 | 29.1 | 29.0 | 33.3 | 29.1 | 29.2 | 33.6 |
| 23 | 34.8 | 34.8 | 34.8/34.6 | 29.6 | 34.8 | 34.6 | 23.7 |
| 24 | 77.5 | 77.6 | 77.5 | 49.4 | 77.7 | 77.7 | 45.0 |
| 25 | 35.8 | 35.8 | 35.9 | 17.8 | 36.1 | 36.2 | 27.8 |
| 26 | 16.4 | 16.4 | 16.4 | 147.5 | 16.5 | 16.5 | 18.3 |
| 27 | 17.5 | 17.5 | 17.5 | 111.3 | 17.6 | 17.6 | 18.3 |
| 28 | 142.1 | 142.3 | 142.2/142.1 | 26.4 | 142.4 | 142.5 | 22.0 |
| 29 | 112.7 | 112.8 | 112.7/112.6 | 11.9 | 112.9 | 112.8 | 11.8 |
| Compounds | Inhibitory Ratio (%) |
|---|---|
| 1 | 3.31 ± 0.85 |
| 2 | 4.08 ± 0.39 |
| 3 | 2.87 ± 0.62 |
| 4 | 8.13 ± 1.76 |
| 5 | 31.28 ± 1.04 |
| 6 | N.I. 1 |
| Isophitol | 21.86 ± 1.21 |
| Indole-3-carboxylic acid | 10.74 ± 0.92 |
| 1-O-Palmitoyl-3-O-(6′-sulfo-α-d-quinovopyranosyl) glycerol | 27.41 ± 1.11 |
| (2S)-1-O-Palmitoyl-3-O-[α-d-galactopyranosyl(1→2)β-d-galactopyranosyl] glycerol | 33. 89 ± 1.03 |
| 3-Methylsulfoxypropionic acid | 12.42 ± 0.63 |
| Tyrosol | 15.81 ± 1.16 |
| 4-Hydroxybenzoic acid | 27.80 ± 0.79 |
| 4-Hydroxyphenylacetic acid | 33.05 ± 1.32 |
| Loliolide | 8.46 ± 1.15 |
| Annuionone D | 18.74 ± 0.92 |
| Azelaic acid | 13.38 ± 0.59 |
| Succinic acid | 15.98 ± 0.87 |
| 8-Hydroxy-(6E)-octenoic acid | 28.92 ± 0.53 |
| n-Butyl β-d-fructopyranoside | 6.41 ± 0.88 |
| n-Butyl pyroglutamate | 16.38 ± 1.87 |
| Quercetin | 71.66 ± 0.52 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.-L.; Guo, W.-J.; Wang, G.-B.; Wang, R.-R.; Hou, Y.-X.; Liu, K.; Liu, Y.; Wang, W. Sterols from the Green Alga Ulva australis. Mar. Drugs 2017, 15, 299. https://doi.org/10.3390/md15100299
Li G-L, Guo W-J, Wang G-B, Wang R-R, Hou Y-X, Liu K, Liu Y, Wang W. Sterols from the Green Alga Ulva australis. Marine Drugs. 2017; 15(10):299. https://doi.org/10.3390/md15100299
Chicago/Turabian StyleLi, Guo-Liang, Wei-Jie Guo, Guang-Bao Wang, Rong-Rong Wang, Yu-Xue Hou, Kun Liu, Yang Liu, and Wei Wang. 2017. "Sterols from the Green Alga Ulva australis" Marine Drugs 15, no. 10: 299. https://doi.org/10.3390/md15100299
APA StyleLi, G. -L., Guo, W. -J., Wang, G. -B., Wang, R. -R., Hou, Y. -X., Liu, K., Liu, Y., & Wang, W. (2017). Sterols from the Green Alga Ulva australis. Marine Drugs, 15(10), 299. https://doi.org/10.3390/md15100299