Collismycin C from the Micronesian Marine Bacterium Streptomyces sp. MC025 Inhibits Staphylococcus aureus Biofilm Formation
Abstract
:1. Introduction
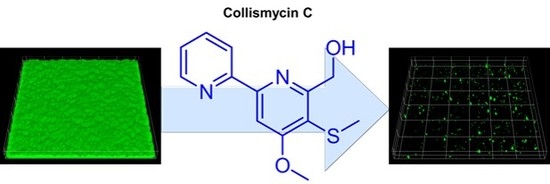
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Isolation of Microbial Strains from Micronesian Marine Samples
3.3. Small-Scale Fermentation and Extraction
3.4. Isolation of 1–6 from Large-Scale Culture Broth of Streptomyces sp. MC025
3.5. Biofilm-Forming Bacterial Strains and Culture Conditions
3.6. Antibiofilm Assays
3.7. Confocal Laser Microscopy
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ventola, C.L. The antibiotic resistance crisis: Part 2: Management strategies and new agents. P & T 2015, 40, 344–352. [Google Scholar]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [PubMed]
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 2005, 436, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Linares, J.F.; Gustafsson, I.; Baquero, F.; Martinez, J.L. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. USA 2006, 103, 19484–19489. [Google Scholar] [PubMed]
- Kaplan, J.B.; Izano, E.A.; Gopal, P.; Karwacki, M.T.; Kim, S.; Bose, J.L.; Bayles, K.W.; Horswill, A.R. Low Levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. MBio 2012, 3, e00198-12. [Google Scholar] [CrossRef] [PubMed]
- San, T.; Ertugay, O.C.; Catli, T.; Acar, M.; Ertugay, C.K.; Dag, I.; Cingi, C. Effects of surfactant on biofilm formation on silicone nasal splints. Eur. Arch. Otorhinolaryngol. 2015, 272, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.J.; Yan, X.; Wu, M.X.; Li, W.K.; Deng, L. Development of an aptamer-ampicillin conjugate for treating biofilms. Biochem. Biophy. Res. Commun. 2017, 483, 847–854. [Google Scholar]
- Yasuda, H.; Ajiki, Y.; Koga, T.; Kawada, H.; Yokota, T. Interaction between biofilms formed by Pseudomonas aeruginosa and clarithromycin. Antimicrob. Agents Chemother. 1993, 37, 1749–1755. [Google Scholar] [PubMed]
- Ren, D.C.; Sims, J.J.; Wood, T.K. Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ. Microbiol. 2001, 3, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Pitts, B.; Stewart, P.S.; Camper, A.; Yoon, J. Comparison of the antimicrobial effects of chlorine, silver ion, and tobramycin on biofilm. Antimicrob. Agents Chemother. 2008, 52, 1446–1453. [Google Scholar] [CrossRef] [PubMed]
- Gomi, S.; Amano, S.; Sato, E.; Miyadoh, S.; Kodama, Y. Novel antibiotics SF2738A, B and C, and their analogs produced by Streptomyces sp. J. Antibiot. 1994, 47, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, N.; Furihata, K.; Shin-Ya, K.; Hayakawa, Y.; Seto, H. Novel Antibiotics pyrisulfoxin A and B produced by Streptomyces californicus. J. Antibiot. 1999, 52, 505–507. [Google Scholar] [CrossRef] [PubMed]
- Funk, A.; Divekar, P.V. Caerulomycin, a new antibiotic from Streptomyces caeruleus Baldacci: I. production, isolation, assay, and biological properties. Can. J. Microbiol. 1959, 5, 317–321. [Google Scholar] [CrossRef] [PubMed]
- McInnes, A.; Smith, D.; Wright, J.; Vining, L. Caerulomycins B and C, new 2,2′-dipyridyl derivatives from Streptomyces caeruleus. Can. J. Chem. 1977, 55, 4159–4165. [Google Scholar] [CrossRef]
- McInnes, A.; Smith, D.; Walter, J.; Wright, J.; Vining, L.; Arsenault, G. Caerulomycin D, a novel glycosidic derivative of 3,4-dihydroxy-2,2′-dipyridyl 6-aldoxime from Streptomyces caeruleus. Can. J. Chem. 1978, 56, 1836–1842. [Google Scholar] [CrossRef]
- Fu, P.; Wang, S.; Hong, K.; Li, X.; Liu, P.; Wang, Y.; Zhu, W. Cytotoxic bipyridines from the marine-derived actinomycete Actinoalloteichus cyanogriseus WH1-2216-6. J. Nat. Prod. 2011, 74, 1751–1756. [Google Scholar] [CrossRef] [PubMed]
- Shindo, K.; Yamagishi, Y.; Okada, Y.; Kawai, H. Collismycins A and B, novel non-steroidal inhibitors of dexamethasone-glucocorticoid receptor binding. J. Antibiot. 1994, 47, 1072–1074. [Google Scholar] [CrossRef] [PubMed]
- Kujur, W.; Gurram, R.K.; Haleem, N.; Maurya, S.K.; Agrewala, J.N. Caerulomycin A inhibits Th2 cell activity: A possible role in the management of asthma. Sci. Rep. 2015, 5, 15396. [Google Scholar] [CrossRef] [PubMed]
- Kujur, W.; Gurram, R.K.; Maurya, S.K.; Nadeem, S.; Chodisetti, S.B.; Khan, N.; Agrewala, J.N. Caerulomycin A suppresses the differentiation of naïve T cells and alleviates the symptoms of experimental autoimmune encephalomyelitis. Autoimmunity 2017, 50, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Kaes, C.; Katz, A.; Hosseini, M.W. Bipyridine: The most widely used ligand. A review of molecules comprising at least two 2,2′-bipyridine units. Chem. Rev. 2000, 100, 3553–3590. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.H.; Shu, J.C.; Huang, H.Y.; Cheng, Y.C. Involvement of iron in biofilm formation by Staphylococcus aureus. PLoS ONE 2012, 7, e34388. [Google Scholar] [CrossRef] [PubMed]
- O’May, C.Y.; Sanderson, K.; Roddam, L.F.; Kirov, S.M.; Reid, D.W. Iron-binding compounds impair Pseudomonas aeruginosa biofilm formation, especially under anaerobic conditions. J. Med. Micriobiol. 2009, 58, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Kawatani, M.; Muroi, M.; Wada, A.; Inoue, G.; Futamura, Y.; Aono, H.; Shimizu, K.; Shimizu, T.; Igarashi, Y.; Takahashi-Ando, N.; et al. Proteomic profiling reveals that collismycin A is an iron chelator. Sci. Rep. 2016, 6, 38385. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef] [PubMed]






© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.-H.; Kim, E.; Choi, H.; Lee, J. Collismycin C from the Micronesian Marine Bacterium Streptomyces sp. MC025 Inhibits Staphylococcus aureus Biofilm Formation. Mar. Drugs 2017, 15, 387. https://doi.org/10.3390/md15120387
Lee J-H, Kim E, Choi H, Lee J. Collismycin C from the Micronesian Marine Bacterium Streptomyces sp. MC025 Inhibits Staphylococcus aureus Biofilm Formation. Marine Drugs. 2017; 15(12):387. https://doi.org/10.3390/md15120387
Chicago/Turabian StyleLee, Jin-Hyung, Eonmi Kim, Hyukjae Choi, and Jintae Lee. 2017. "Collismycin C from the Micronesian Marine Bacterium Streptomyces sp. MC025 Inhibits Staphylococcus aureus Biofilm Formation" Marine Drugs 15, no. 12: 387. https://doi.org/10.3390/md15120387
APA StyleLee, J. -H., Kim, E., Choi, H., & Lee, J. (2017). Collismycin C from the Micronesian Marine Bacterium Streptomyces sp. MC025 Inhibits Staphylococcus aureus Biofilm Formation. Marine Drugs, 15(12), 387. https://doi.org/10.3390/md15120387