Antibacterial Activity of AI-Hemocidin 2, a Novel N-Terminal Peptide of Hemoglobin Purified from Arca inflata
Abstract
:1. Introduction
2. Results and Discussion
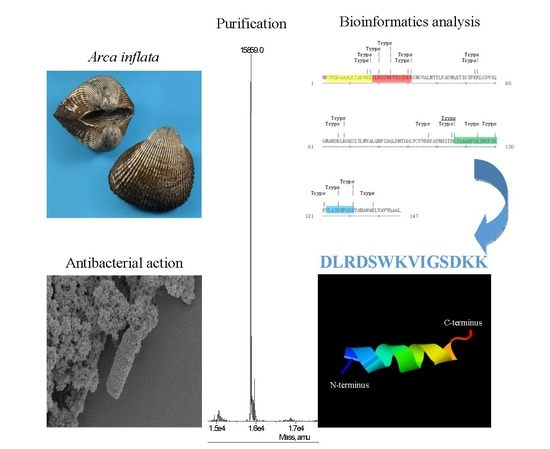
2.1. Purification of Hb-I
2.2. Identification of Hb-I
2.3. Bioinformatic Analysis and Antibacterial Function Screening
2.4. Circular Dichroism Spectra and 3D Secondary Structure Prediction
2.5. Assays for Hemolysis and Cytotoxicity
2.6. Effects of AI-Hemocidin 2 on the Permeability of the Bacterial Membrane
2.7. Propidium Iodide Uptake Experiment
2.8. Examination of the Morphologic Changes in Cells by Scanning Electron Microscopy
3. Materials and Methods
3.1. Materials and Strain Culture Conditions
3.2. Screening for Antibacterial Activity Components
3.3. Purification of Hemoglobin Hb-I
3.4. Molecular Mass and Amino Acid Sequence Determination of Hemoglobin Hb-I
3.5. Bioinformatics Analysis
3.6. Peptide Synthesis
3.7. Determination of the Antibacterial Spectrum and Minimum Inhibitory Concentration
3.8. Hemolytic Assay and Cell Viability
3.9. Effects of pH and Temperature on the Antibacterial Activity
3.10. Membrane Permeability Assay
3.11. Integrity of the Bacterial Membrane
3.12. Examination of Bacterial Membrane Damage by SEM
3.13. CD Determination and Secondary Structure Prediction
3.14. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Authors Contributions
Conflicts of Interest
References
- Centers for Disease Control and Prevention, Office of Infectious Disease. Antibiotic Resistance Threats in the United States. Available online: http://www.cdc.gov/drugresistance/threat-report-2013 (accessed on 12 April 2013).
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 2014, 8, 129–136. [Google Scholar] [PubMed]
- Gould, I.M.; Bal, A.M. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence 2013, 4, 185–191. [Google Scholar] [PubMed]
- Congressional Research Service Report. Life Expectancy in the United States. Available online: http://www.menshealthnetwork.org/library/CRSlifeexpectRL32792.pdf (accessed on 12 April 2017).
- Sengupta, S.; Chattopadhyay, M.K.; Grossart, H.P. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 2013, 4, 1–13. [Google Scholar]
- Wright, G.D. Something old, something new: Revisiting natural products in antibiotic drug discovery. Can. J. Microbiol. 2014, 60, 147–154. [Google Scholar] [PubMed]
- Brogan, D.M.; Mossialos, E. A critical analysis of the review on antimicrobial resistance report and the infectious disease financing facility. Glob. Health 2016, 12, 1–7. [Google Scholar]
- Lin, M.C.; Lin, S.B.; Lee, S.C.; Lin, C.C.; Hui, C.F.; Chen, J.Y. Antimicrobial peptide of an anti-lipopolysaccharide factor modulates of the inflammatory response in raw264.7 cells. Peptides 2010, 31, 1262–1272. [Google Scholar] [PubMed]
- Stark, M.; Liu, L.P.; Deber, C.M. Cationic hydrophobic peptides with antimicrobial activity. Antimicrob. Agents Chemother. 2002, 46, 3585–3590. [Google Scholar] [PubMed]
- Van ’t Hof, W.; Veerman, E.C.I.; Helmerhorst, E.J.; Amerongen, A.V.N. Antimicrobial peptides properties and applicability. Biol. Chem. 2001, 382, 597–619. [Google Scholar] [PubMed]
- Izadpanah, A.; Gallo, R.L. Antimicrobial peptides. J. Am. Acad. Dermatol. 2005, 52, 381–390. [Google Scholar] [PubMed]
- Wimley, W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Aneiros, A.; Garateix, A. Bioactive peptides from marine sources: Pharmacological properties and isolation procedures. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2004, 803, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Harnedy, P.A.; FitzGerald, J.F. Bioactive peptides from marine processing waste and shellfish: A review. J. Funct. Foods 2012, 4, 6–24. [Google Scholar] [CrossRef]
- Nam, B.H.; Moon, J.Y.; Park, E.H.; Kong, H.J.; Kim, Y.O.; Kim, D.G.; Kim, W.J.; An, C.M.; Seo, J.K. Antimicrobial and antitumor activities of novel peptides derived from the lipopolysaccharide- and-1,3-glucan binding protein of the pacific abalone haliotis discus hannai. Mar. Drugs 2016, 14, 227. [Google Scholar] [CrossRef] [PubMed]
- Simon-Colin, C.; Gueguen, Y.; Bachere, E.; Kouzayha, A.; Saulnier, D.; Gayet, N.; Guezennec, J. Use of natural antimicrobial peptides and bacterial biopolymers for cultured pearl production. Mar. Drugs 2015, 13, 3732–3744. [Google Scholar] [CrossRef] [PubMed]
- Parish, C.A.; Jiang, H.; Tokiwa, Y.; Berova, N.; Nakanishi, K.; McCabe, D.; Zuckerman, W.; Xia, M.M.; Gabay, J.E. Broad-spectrum antimicrobial activity of hemoglobin. Bioorgan. Med. Chem. 2001, 9, 377–382. [Google Scholar] [CrossRef]
- Ivanov, V.T.; Karelin, A.A.; Philippova, M.M.; Nazimov, I.V.; Pletnev, V.Z. Hemoglobin as a source of endogenous bioactive peptides: The concept of tissue-specific peptide pool. Biopolymers 1997, 43, 171–188. [Google Scholar] [CrossRef]
- Ullal, A.J.; Litaker, R.W.; Noga, E.J. Antimicrobial peptides derived from hemoglobin are expressed in epithelium of channel catfish (ictalurus punctatus, rafinesque). Dev. Comp. Immunol. 2008, 32, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Fogaca, A.C.; da Silva, P.I.J.; Miranda, M.T.; Bianchi, A.G.; Miranda, A.; Ribolla, P.E.; Daffre, S. Antimicrobial activity of a bovine hemoglobin fragment in the tick boophilus microplus. J. Biol. Chem. 1999, 274, 25330–25334. [Google Scholar] [CrossRef] [PubMed]
- Liepke, C.; Baxmann, S.; Heine, C.; Breithaupt, N.; Ständker, L.; Forssmann, W.G. Human hemoglobin-derived peptides exhibit antimicrobial activity: A class of host defense peptides. J. Chromatogr. B 2003, 791, 345–356. [Google Scholar] [CrossRef]
- Catiau, L.; Traisnel, J.; Chihib, N.E.; Le Flem, G.; Blanpain, A.; Melnyk, O.; Guillochon, D.; Nedjar-Arroume, N.R. A minimal peptidic sequence obtained from beta-chain hemoglobin exhibiting an antimicrobial activity. Peptides 2011, 32, 1463–1468. [Google Scholar] [CrossRef] [PubMed]
- Mak, P.; Wojcik, K.; Wicherek, L.; Suder, P.; Dubin, A. Antibacterial hemoglobin peptides in human menstrual blood. Peptides 2004, 25, 1839–1847. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.H.; Xu, Y.Z.; Wang, Q.; Hang, B.L.; Sun, Y.W.; Wei, X.X.; Hu, J.H. Potential of novel antimicrobial peptide p3 from bovine erythrocytes and its analogs to disrupt bacterial membranes in vitro and display activity against drug-resistant bacteria in a mouse model. Antimicrob. Agents Chemother. 2015, 59, 2835–2841. [Google Scholar] [CrossRef] [PubMed]
- Catiau, L.; Traisnel, J.; Delval-Dubois, V.; Chihib, N.E.; Guillochon, D.; Nedjar-Arroume, N. Minimal antimicrobial peptidic sequence from hemoglobin alpha-chain: Kyr. Peptides 2011, 32, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Coates, C.J.; Decker, H. Immunological properties of oxygen-transport proteins: Hemoglobin, hemocyanin and hemerythrin. Cell. Mol. Life Sci. 2017, 74, 293–317. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Coates, C.J.; Zhu, H.; Zhu, P.; Wu, Z.; Xie, L. Identification of candidate antimicrobial peptides derived from abalone hemocyanin. Dev. Comp. Immunol. 2015, 49, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chen, Z.; Song, L.; Chen, L.; Zhu, J.; Lv, S.; Yu, R. A new in vitro anti-tumor polypeptide isolated from Arca inflata. Mar. Drugs 2013, 11, 4773–4787. [Google Scholar] [CrossRef] [PubMed]
- Rowley, A.F.; Powell, A. Invertebrate immune systems–specific, quasi-specific, or nonspecific? J. Immunol. 2007, 179, 7209–7214. [Google Scholar] [CrossRef] [PubMed]
- Mine, Y.; Ma, F.P.; Lauriau, S. Antimicrobial peptides released by enzymatic hydrolysis of hen egg white lysozyme. J. Agric. Food Chem. 2004, 52, 1088–1094. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, N.N.; Alam, H.; Pandey, S.; Gautam, L.; Sinha, M.; Shin, K.; Manzoor, N.; Virdi, J.S.; Kaur, P.; Sharma1, S.; et al. Preparation and antimicrobial action of three tryptic digested functional molecules of bovine lactoferrin. PLoS ONE 2014, 9, e90011. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, C.; Zhu, J.; Xie, W.; Hu, X.; Song, L.; Zi, J.; Yu, R. Purification and characterization of an antibacterial and anti-inflammatory polypeptide from arca subcrenata. Int. J. Biol. Macromol. 2017, 96, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhao, H.; Zhang, C.; Yu, J.; Lu, Z. Purification and characterization of plantaricin 163, a novel bacteriocin produced by Lactobacillus plantarum 163 isolated from traditional Chinese fermented vegetables. J. Agric. Food Chem. 2013, 61, 11676–11682. [Google Scholar] [CrossRef] [PubMed]
- Hata, T.; Tanaka, R.; Ohmomo, S. Isolation and characterization of plantaricin ASM1: A new bacteriocin produced by Lactobacillus plantarum A-1. Int. J. Food Microbiol. 2010, 137, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Shi, B.; Zhu, D.; Cai, Q.; Chen, Y.; Li, J.; Qi, K.; Zhang, M. Characterization of a novel bacteriocin produced by Lactobacillus sakei LSJ618 isolated from traditional Chinese fermented radish. Food Control 2012, 23, 338–344. [Google Scholar] [CrossRef]
- Pitout, J.D. Extraintestinal pathogenic Escherichia coli: An update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev. Anti-Infect. Ther. 2012, 10, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Froidevaux, R.; Krier, F.; Nedjar-Arroume, N.; Vercaigne-Marko, D.; Kosciarz, E.; Ruckebusch, C.; Dhulster, P.; Guillochon, D. Antibacterial activity of a pepsin-derived bovine hemoglobin fragment. FEBS Lett. 2001, 491, 159–163. [Google Scholar] [CrossRef]
- Pakdeesuwan, A.; Araki, T.; Daduang, S.; Payoungkiattikun, W.; Jangpromma, N.; Klaynongsruang, S. In vivo wound healing activity of crocodile (crocodylus siamensis) hemoglobin and evaluation of antibacterial and antioxidant properties of hemoglobin and hemoglobin hydrolysate. J. Microbiol. Biotechnol. 2016, 27, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, C.L.; Rozek, A.; Patrzykat, A.; Hancock, R.E.W. Structure and mechanism of action of an indolicidin peptide derivative with improved activity against gram-positive bacteria. J. Biol. Chem. 2001, 276, 24015–24022. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Dathe, M.; Wieprecht, T. Structural features of helical antimicrobial peptides: Their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta 1999, 1462, 71–87. [Google Scholar] [CrossRef]
- Lopez-Abarrategui, C.; McBeth, C.; Mandal, S.M.; Sun, Z.J.; Heffron, G.; Alba-Menendez, A.; Migliolo, L.; Reyes-Acosta, O.; Garcia-Villarino, M.; Nolasco, D.O.; et al. Cm-p5: An antifungal hydrophilic peptide derived from the coastal mollusk cenchritis muricatus (gastropoda: Littorinidae). FASEB J. 2015, 29, 3315–3325. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Yang, Y.; Lyu, X.; Dong, N.; Shan, A. Antimicrobial activity, improved cell selectivity and mode of action of short pmap-36-derived peptides against bacteria and candida. Sci. Rep. 2016, 6, e27258. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lee, M.J.; Jung, H.G.; Go, H.J.; Kim, Y.J.; Park, N.G. Antimicrobial function of SHbetaAP, a novel hemoglobin beta chain-related antimicrobial peptide, isolated from the liver of skipjack tuna, katsuwonus pelamis. Fish Shellfish Immunol. 2014, 37, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Jeong, M.; Jeon, D.; Shin, A.; Jin, S.; Shin, S.; Park, Y.; Kim, Y. Effects of Hydrophobic Peptoid Substitutions on the Bacterial Cell Selectivity and Antimicrobial Activity of Piscidin 1. Bull. Korean Chem. Soc. 2016, 37, 1545–1551. [Google Scholar] [CrossRef]
- Lee, E.; Shin, A.; Jeong, K.W.; Jin, B.; Jnawali, H.N.; Shin, S.; Shin, S.Y.; Kim, Y. Role of phenylalanine and valine (10) residues in the antimicrobial activity and cytotoxicity of piscidin-1. PLoS ONE 2014, 9, e114453. [Google Scholar] [CrossRef] [PubMed]
- Phadke, S.M.; Lazarevic, V.; Bahr, C.C.; Islam, K.; Stolz, D.B.; Watkins, S.; Tencza, S.B.; Vogel, H.J.; Montelaro, R.C.; Mietzner, T.A. Lentivirus lytic peptide 1 perturbs both outer and inner membranes of serratia marcescens. Antimicrob. Agents Chemother. 2002, 46, 2041–2045. [Google Scholar] [CrossRef] [PubMed]
- Skerlavaj, B.; Romeo, D.; Gennaro, R. Rapid membrane permeabilization and inhibition of vital functions of gram-negative bacteria by bactenecins. Infect. Immun. 1990, 58, 3724–3730. [Google Scholar] [PubMed]
- Scocchi, M.; Mardirossian, M.; Runti, G.; Benincasa, M. Non-membrane permeabilizing modes of action of antimicrobial peptides on bacteria. Curr. Top. Med. Chem. 2016, 16, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.K.; Han, E.J.; Han, K.J.; Paik, H.D. Antimicrobial effect of bacteriocin ku24 produced by lactococcus lactis ku24 against methicillin-resistant Staphylococcus aureus. J. Food Sci. 2013, 78, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Piers, K.L.; Brown, M.H.; Hancock, R.E. Improvement of outer membrane-permeabilizing and lipopolysaccharide-binding activities of an antimicrobial cationic peptide by c-terminal modification. Antimicrob. Agents Chemother. 1994, 38, 2311–2316. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Bai, X.; Luan, N.; Yao, H.; Zhang, Z.; Liu, W.; Chen, Y.; Yan, X.; Rong, M.; Lai, R. A designed tryptophan- and lysine/arginine-rich antimicrobial peptide with therapeutic potential for clinical antibiotic-resistant candida albicans vaginitis. J. Med. Chem. 2016, 59, 1791–1799. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lopez, S.; Choi, E.C.; Delgado, M.; Granja, J.R.; Khasanov, A.; Kraehenbuehl, K.; Long, G.; Weinberger, D.A.; Wilcoxen, K.M.; Ghadiri, M.R. Antibacterial agents based on the cyclic d,l-alpha-peptide architecture. Nature 2001, 412, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Adje, E.Y.; Balti, R.; Kouach, M.; Dhulster, P.; Guillochon, D.; Nedjar-Arroume, N. Obtaining antimicrobial peptides by controlled peptic hydrolysis of bovine hemoglobin. Int. J. Biol. Macromol. 2011, 49, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Wang, J.; Gao, H.; Wang, Z.; Dong, N.; Ma, Q.; Shan, A. Antimicrobial properties and membrane-active mechanism of a potential alpha-helical antimicrobial derived from cathelicidin pmap-36. PLoS ONE 2014, 9, e86364. [Google Scholar]
- Miao, J.; Guo, H.; Chen, F.; Zhao, L.; He, L.; Ou, Y.; Huang, M.; Zhang, Y.; Guo, B.; Cao, Y. Antibacterial effects of a cell-penetrating peptide isolated from kefir. J. Agric. Food Chem. 2016, 64, 3234–3242. [Google Scholar] [CrossRef] [PubMed]
- Schaller, A.; Sun, Z.H.; Yang, Y.P.; Somoskovi, A.; Zhang, Y. Salicylate reduces susceptibility of mycobacterium tuberculosis to multiple antituberculosis drugs. Antimicrob. Agents Chemother. 2002, 46, 2636–2639. [Google Scholar] [CrossRef] [PubMed]








| Fractions | J1 | J2 | J3 | J4 | J1S1 | J1S2 | J1S3 | Cipro |
|---|---|---|---|---|---|---|---|---|
| IZ (mm) | 10.18 ± 0.24 | 9.22 ± 0.21 | nd | nd | 16.19 ± 0.62 | nd | 6.74 ± 0.18 | 31.66 ± 0.22 |
| Peptide | Sequence | Length | pI | Mw | NC | H | PR/n % |
|---|---|---|---|---|---|---|---|
| 1 | PSVQGAAAQLTADVKK | 16 | 9.01 | 1583.81 | 1 | 0.196 | 8/50% |
| 2 | DLRDSWKVIGSDKK | 14 | 8.50 | 1646.86 | 1 | 0.043 | 10/71.43% |
| 3 | ISAAEFGKINGPIKK | 15 | 9.70 | 1572.87 | 2 | 0.285 | 8/53.33% |
| 4 | VLASKNFGDK | 10 | 8.56 | 1078.23 | 1 | 0.163 | 6/60% |
| Bacteria | MIC Value (μM) | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | Hb-I | Cipro | |
| E. coli ATCC 25922 | 47.35 | 45.54 | >381.47 | >556.47 | 75.66 | 24.14 |
| P. aeruginosa ATCC 27853 | 47.35 | 91.08 | 95.37 | >556.47 | >100.88 | 48.28 |
| P. aeruginosa clinical isolate | 47.35 | 91.08 | >381.47 | >556.47 | >100.88 | 48.28 |
| S. aureus ATCC 25923 | >378.83 | 22.77 | >381.47 | >556.47 | 75.66 | 24.14 |
| B. subtilis ATCC 6633 | >378.83 | 182.16 | >381.47 | 69.55 | >100.88 | 12.07 |
| B. subtilis ATCC 6633 | >378.83 | 182.16 | >381.47 | >556.47 | >100.88 | 12.07 |
| E. faecalis ATCC 29212 | >378.83 | >364.33 | >381.47 | >556.47 | >100.88 | 24.14 |
| Sample | AI-Hemocidin-2 |
|---|---|
| MIC a (μg/mL) | 37.5–300 |
| HC10 b (μg/mL) | >500 |
| Hmax c | 10.27 ± 0.42 |
| Cell viability (%) at 250 μg/mL | 88.18 ± 9.48 |
| IC50 d(μg/mL) | >1000 |
| SI value e | 3.33–26.67 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Zhu, J.; Wang, Y.; Chen, Y.; Song, L.; Zheng, W.; Li, J.; Yu, R. Antibacterial Activity of AI-Hemocidin 2, a Novel N-Terminal Peptide of Hemoglobin Purified from Arca inflata. Mar. Drugs 2017, 15, 205. https://doi.org/10.3390/md15070205
Li C, Zhu J, Wang Y, Chen Y, Song L, Zheng W, Li J, Yu R. Antibacterial Activity of AI-Hemocidin 2, a Novel N-Terminal Peptide of Hemoglobin Purified from Arca inflata. Marine Drugs. 2017; 15(7):205. https://doi.org/10.3390/md15070205
Chicago/Turabian StyleLi, Chunlei, Jianhua Zhu, Yanqing Wang, Yuyan Chen, Liyan Song, Weiming Zheng, Jingjing Li, and Rongmin Yu. 2017. "Antibacterial Activity of AI-Hemocidin 2, a Novel N-Terminal Peptide of Hemoglobin Purified from Arca inflata" Marine Drugs 15, no. 7: 205. https://doi.org/10.3390/md15070205
APA StyleLi, C., Zhu, J., Wang, Y., Chen, Y., Song, L., Zheng, W., Li, J., & Yu, R. (2017). Antibacterial Activity of AI-Hemocidin 2, a Novel N-Terminal Peptide of Hemoglobin Purified from Arca inflata. Marine Drugs, 15(7), 205. https://doi.org/10.3390/md15070205