Sterols from the Octocoral Nephthea columnaris
Abstract
:1. Introduction
2. Results and Discussion
3. Experimental Section
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Extraction and Isolation
3.4. Molecular Mechanics Calculations
3.5. Generation of Superoxide Anions and Release of Elastase by Human Neutrophils
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hsiao, T.-H.; Sung, C.-S.; Lan, Y.-H.; Wang, Y.-C.; Lu, M.-C.; Wen, Z.-H.; Wu, Y.-C.; Sung, P.-J. New anti- inflammatory cembranes from the cultured soft coral Nephthea columnaris. Mar. Drugs 2015, 13, 3443–3453. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, T.-H.; Cheng, C.-H.; Wu, T.-Y.; Lu, M.-C.; Chen, W.-F.; Wen, Z.-H.; Dai, C.-F.; Wu, Y.-C.; Sung, P.-J. New cembranoid diterpenes from the cultured Nephthea columnaris. Molecules 2015, 20, 13205–13215. [Google Scholar] [CrossRef] [PubMed]
- Whuang, T.-Y.; Tsai, W.-C.; Chen, N.-F.; Chen, Z.-C.; Tsui, K.-H.; Wen, Z.-H.; Su, Y.-D.; Chang, Y.-C.; Chen, Y.-H.; Lu, M.-C.; et al. Columnaristerol A, a novel 19-norsterol from the Formosan octocoral Nephthea columnaris. Bioorg. Med. Chem. Lett. 2016, 26, 4966–4969. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-C.; Whuang, T.-Y.; Su, J.-H.; Hwang, T.-L.; Wu, Y.-C.; Sung, P.-J. 4α-Methylergosta-22(E), 24(28)-dien-3β-ol, a new marine sterol from the octocoral Nephthea columnaris. Nat. Prod. Commun. 2017, 12, 345–346. [Google Scholar]
- Iguchi, K.; Saitoh, S.; Yamada, Y. Novel 19-oxygenated sterols from the Okinawan soft coral Litophyton viridis. Chem. Pharm. Bull. 1989, 37, 2553–2554. [Google Scholar] [CrossRef]
- McInnes, A.G.; Walter, J.A.; Wright, J.L.C. 13C NMR spectra of Δ24(28) phytosterols. Org. Magn. Reson. 1980, 13, 302–303. [Google Scholar] [CrossRef]
- Allinger, N.L. Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. J. Am. Chem. Soc. 1977, 99, 8127–8134. [Google Scholar] [CrossRef]
- Van Ofwegen, L.P. Annotated check list of New Caledonian soft corals. In Compendium of Marine Species of New Caledonia, 2nd ed.; Payri, C.E., Richer de Forges, B., Eds.; Institut de Recherche Pour le Développement: Nouméa, New Caledonia, France, 2007; pp. 139–144. [Google Scholar]
- Yang, S.-C.; Chung, P.-J.; Ho, C.-M.; Kuo, C.-Y.; Hung, M.-F.; Huang, Y.-T.; Chang, W.-Y.; Chang, Y.-W.; Chan, K.-H.; Hwang, T.-L. Propofol inhibits superoxide production, elastase release, and chemotaxis in formyl peptide-activated human neutrophils by blocking formyl peptide receptor 1. J. Immunol. 2013, 190, 6511–6519. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-P.; Hsieh, P.-W.; Chang, Y.-J.; Chung, P.-J.; Kuo, L.-M.; Hwang, T.-L. 2-(2-Fluorobenzamido) benzoate ethyl ester (EFB-1) inhibits superoxide production by human neutrophils and attenuates hemorrhagic shock-induced organ dysfunction in rats. Free Radic. Biol. Med. 2011, 50, 1737–1748. [Google Scholar] [CrossRef] [PubMed]



| Position | δH (J in Hz) | δC, Multiple | 1H–1H COSY | HMBC |
|---|---|---|---|---|
| 1α/β | 1.84 m; 1.09 m | 37.2, CH2 | H2-2 | C-2, C-3, C-5, C-10, C-19 |
| 2α/β | 1.84 m; 1.49 m | 31.6, CH2 | H2-1, H-3 | C-3 |
| 3 | 3.51 m | 71.7, CH | H2-2, H2-4 | n. o. a |
| 4a/b | 2.31 ddd (12.8, 4.8, 2.0); 2.22 m | 42.1, CH2 | H-3 | C-2, C-3, C-5, C-6, C-10 |
| 5 | - | 140.6, C | - | - |
| 6 | 5.35 br d (5.2) | 121.6, CH | H2-7 | C-4, C-7, C-8, C-10 |
| 7a/b | 1.99 m; 1.49 m | 31.5, CH2 | H-6, H-8 | C-5, C-6, C-8, C-9 |
| 8 | 1.36 m | 30.6, CH | H2-7, H-9, H-14 | C-7, C-9, C-10, C-13, C-14 |
| 9 | 1.06 m | 49.5, CH | H-8, H2-11 | C-7, C-11 |
| 10 | - | 36.5, C | - | - |
| 11a/b | 1.71 m; 1.49 m | 31.3, CH2 | H-9, H-12 | C-8, C-9, C-12, C-13 |
| 12 | 3.46 ddd (10.8, 5.2, 4.4) | 79.5, CH | H2-11 | C-18 |
| 13 | - | 47.6, C | - | - |
| 14 | 0.92 m | 54.7, CH | H-8, H2-15 | C-8, C-12, C-13, C-15, C-18 |
| 15a/b | 1.64 m; 1.21 m | 23.8, CH2 | H-14, H2-16 | C-8, C-14, C-16, C-17 |
| 16a/b | 1.76 m; 1.52 m | 24.4, CH2 | H2-15, H-17 | n. o. |
| 17 | 1.44 m | 57.3, CH | H2-16, H-20 | C-12, C-13, C-16, C-18, C-20, C-21, C-22 |
| 18 | 0.72 s | 7.8, CH3 | - | C-12, C-13, C-14, C-17 |
| 19 | 1.02 s | 19.3, CH3 | - | C-1, C-5, C-9, C-10 |
| 20 | 1.77 m | 33.4, CH | H-17, H3-21, H2-22 | C-16 |
| 21 | 1.04 d (6.4) | 21.5, CH3 | H-20 | C-17 |
| 22a/b | 1.64 m; 1.14 m | 33.4, CH2 | H-20, H2-23 | C-21 |
| 23a/b | 2.12 m; 1.92 m | 32.5, CH2 | H2-22 | C-20, C-22, C-24, C-28 |
| 24 | - | 156.8, C | - | - |
| 25 | 2.23 m | 33.8, CH | H3-26, H3-27 | C-23, C-24, C-26, C-27, C-28 |
| 26 | 1.02 d (6.8) | 21.8, CH3 | H-25 | C-24, C-25, C-27 |
| 27 | 1.03 d (6.8) | 22.0, CH3 | H-25 | C-24, C-25, C-26 |
| 28a/b | 4.72 br s; 4.67 br s | 106.0, CH2 | - | C-23, C-24, C-25 |
| Position | δH (J in Hz) | δC, Multiple | 1H–1H COSY | HMBC |
|---|---|---|---|---|
| 1a/b | 1.81 m, 1.01 m | 37.2, CH2 | H2-2 | C-5 |
| 2a/b | 1.83 m, 1.48 m | 31.6, CH2 | H2-1, H-3 | n. o. a |
| 3 | 3.53 m | 71.7, CH | H2-2, H2-4 | n. o. |
| 4α/β | 2.32 m, 2.21 m | 42.2, CH2 | H-3 | C-3, C-5, C-6 |
| 5 | - | 140.9, C | - | - |
| 6 | 5.38 m | 121.2, CH | H2-7 | C-4, C-7, C-8, C-10 |
| 7a/b | 2.30 m, 1.69 m | 31.0, CH2 | H-6, H-8 | C-5, C-6 |
| 8 | 1.94 m | 27.6, CH | H2-7, H-9, H-14 | C-14 |
| 9 | 1.08 m | 50.5, CH | H-8, H2-11 | C-19 |
| 10 | - | 36.6, C | - | - |
| 11 | 1.50 m | 21.4, CH2 | H-9, H2-12 | n. o. |
| 12a/b | 2.12 m, 1.17 m | 38.9, CH2 | H2-11 | C-18 |
| 13 | - | 46.4, C | - | - |
| 14 | 1.14 m | 60.4, CH | H-8, H2-15 | C-8, C-9, C-12, C-13, C-16, C-17, C-18 |
| 15a/b | 2.42 m, 1.50 m | 41.8, CH2 | H-14, H-16 | C-13, C-16, C-17 |
| 16 | 4.25 m | 70.1, CH | H2-15, H-17 | C-13, C-17 |
| 17 | 1.20 m | 55.8, CH | H-16, H-20 | C-13, C-15, C-18, C-20 |
| 18a/b | 4.00 d (12.0), 3.68 d (12.0) | 62.8, CH2 | - | C-12, C-13, C-14, C-17 |
| 19 | 1.03 s | 19.4, CH3 | - | C-1, C-5, C-9, C-10 |
| 20 | 1.95 m | 34.9, CH | H-17, H3-21, H2-22 | C-22 |
| 21 | 1.04 d (6.4) | 19.2, CH3 | H-20 | C-17, C-20, C-22 |
| 22a/b | 1.55 m, 1.17 m | 34.9, CH2 | H-20, H2-23 | n. o. |
| 23a/b | 2.10 m, 1.91 m | 30.7, CH2 | H2-22 | C-20, C-22, C-24, C-28 |
| 24 | - | 156.6, C | - | - |
| 25 | 2.21 m | 33.8, CH | H3-26, H3-27 | C-23, C-24, C-26, C-27, C-28 |
| 26 | 1.02 d (6.8) | 21.9, CH3 | H-25 | C-24, C-27 |
| 27 | 1.02 d (6.8) | 22.0, CH3 | H-25 | C-24, C-26 |
| 28a/b | 4.72 br s, 4.66 br s | 106.0, CH2 | - | C-23, C-25 |
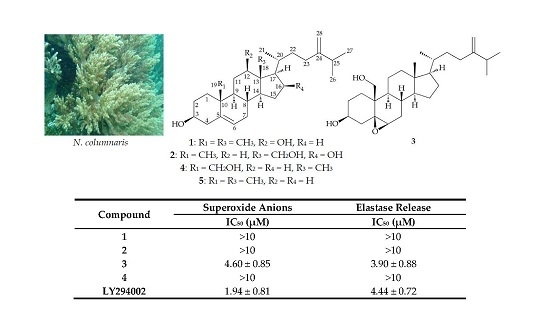
| Compound | Superoxide Anions | Elastase Release |
|---|---|---|
| IC50 (μM) a | IC50 (μM) | |
| 1 | >10 | >10 |
| 2 | >10 | >10 |
| 3 | 4.60 ± 0.85 | 3.90 ± 0.88 |
| 4 | >10 | >10 |
| LY294002 b | 1.94 ± 0.81 | 4.44 ± 0.72 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Whuang, T.-Y.; Tsai, H.-C.; Su, Y.-D.; Hwang, T.-L.; Sung, P.-J. Sterols from the Octocoral Nephthea columnaris. Mar. Drugs 2017, 15, 212. https://doi.org/10.3390/md15070212
Whuang T-Y, Tsai H-C, Su Y-D, Hwang T-L, Sung P-J. Sterols from the Octocoral Nephthea columnaris. Marine Drugs. 2017; 15(7):212. https://doi.org/10.3390/md15070212
Chicago/Turabian StyleWhuang, Ta-Yuan, Hong-Chieh Tsai, Yin-Di Su, Tsong-Long Hwang, and Ping-Jyun Sung. 2017. "Sterols from the Octocoral Nephthea columnaris" Marine Drugs 15, no. 7: 212. https://doi.org/10.3390/md15070212
APA StyleWhuang, T. -Y., Tsai, H. -C., Su, Y. -D., Hwang, T. -L., & Sung, P. -J. (2017). Sterols from the Octocoral Nephthea columnaris. Marine Drugs, 15(7), 212. https://doi.org/10.3390/md15070212