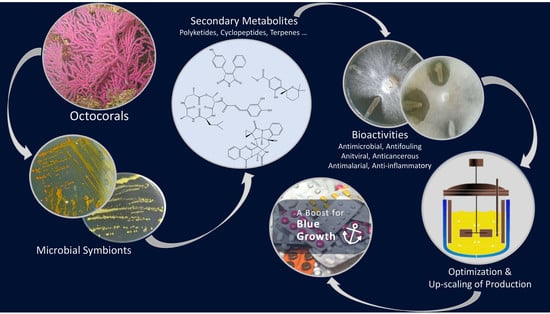
Bioactive Secondary Metabolites from Octocoral-Associated Microbes—New Chances for Blue Growth
Abstract
:1. Introduction
2. Diversity and Function of the Octocoral Microbiome
3. Octocoral-Associated Microbes as Natural-Product Manufacturers
3.1. Antibacterial and Antifungal Activity
3.2. Antifouling Activity
3.3. Antiviral Activity
3.4. Anticancer Activity
3.5. Antimalarial, Anti-Inflammatory, Antineurodegenerative and Other Activities
4. Genomic Insights into Natural Product Biosynthesis by Bacterial Symbionts of Octocorals
Genomic Insights into Biocatalysts
5. Optimization of Bioactive Compound Production to Meet Industrial Demands
6. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munroa, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 33, 382–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Look, S.A.; Fenical, W.; Robert, S.J.; Clardy, J. The Pseudopterosins: Anti-inflammatory and analgesic natural products from the sea whip Pseudopterogorgia elisabethae. Proc. Natl. Acad. Sci. USA 1986, 83, 6238–6240. [Google Scholar] [CrossRef] [PubMed]
- Ospina, C.A.; Rodríguez, A.D.; Zhao, H.; Raptis, R.G. Bipinnapterolide B. a bioactive oxapolycyclic diterpene from the Colombian gorgonian coral Pseudopterogorgia bipinnata. Tetrahedron Lett. 2007, 48, 7520–7523. [Google Scholar] [CrossRef]
- Ospina, C.A.; Rodríguez, A.D.; Sánchez, J.A.; Ortega-Barria, E.; Capson, T.L.; Mayer, A.M. Caucanolides A−F, unusual antiplasmodial constituents from a Colombian collection of the gorgonian coral Pseudopterogorgia bipinnata. J. Nat. Prod. 2005, 68, 1519–1526. [Google Scholar] [CrossRef] [PubMed]
- Berrue, F.; Kerr, R.G. Diterpenes from gorgonian corals. Nat. Prod. Rep. 2009, 26, 681–710. [Google Scholar] [CrossRef] [PubMed]
- Sheu, J.H.; Chen, Y.H.; Chen, Y.H.; Su, Y.D.; Chang, Y.C.; Su, J.H.; Weng, C.F.; Lee, C.H.; Fang, L.S.; Wang, W.H.; et al. Briarane diterpenoids isolated from gorgonian corals between 2011 and 2013. Mar. Drugs 2014, 12, 2164–2181. [Google Scholar] [CrossRef] [PubMed]
- Rocha, J.; Peixe, L.; Gomes, N.C.M.; Calado, R. Cnidarians as a source of new marine bioactive compounds—An overview of the last decade and future steps for bioprospecting. Mar. Drugs 2011, 9, 1860–1886. [Google Scholar] [CrossRef]
- Changyun, W.; Haiyan, L.; Changlun, S.; Yanan, W.; Liang, L.; Huashi, G. Chemical defensive substances of soft corals and gorgonians. Acta Ecol. Sin. 2008, 28, 2320–2328. [Google Scholar]
- Berrue, F.; Withers, S.T.; Haltli, B.; Withers, J.; Kerr, R.G. Chemical screening method for the rapid identification of microbial sources of marine invertebrate-associated metabolites. Mar. Drugs 2011, 9, 369–381. [Google Scholar] [CrossRef]
- Bhanot, A.; Sharma, R.; Noolvi, M.N. Natural sources as potential anti-cancer agents: A review. Int. J. Phytomed. 2011, 3, 9–26. [Google Scholar]
- Piel, J.; Hui, D.Q.; Wen, G.P.; Butzke, D.; Platzer, M.; Fusetani, N.; Matsunaga, S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 2004, 101, 16222–16227. [Google Scholar] [CrossRef] [PubMed]
- Davidson, S.K.; Allen, S.W.; Lim, G.E.; Anderson, C.M.; Haygood, M.G. Evidence for the biosynthesis of Bryostatins by the bacterial symbiont “Candidatus Endobugula sertula” of the bryozoan Bugula neritina. Appl. Environ. Microbiol. 2001, 67, 4531–4537. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Kuzuyama, T.; Komatsu, M.; Shin-ya, K.; Omura, S.; Cane, D.E.; Ikeda, H. Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Piel, J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 2002, 99, 14002–14007. [Google Scholar] [CrossRef] [PubMed]
- Piel, J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2004, 21, 519–538. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.C.; Mori, T.; Rückert, C.; Uria, A.R.; Helf, M.J.; Takada, K.; Gernert, C.; Steffens, U.A.; Heycke, N.; Schmitt, S.; et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 2014, 506, 58–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebel, R. Terpenes from marine-derived fungi. Mar. Drugs 2010, 8, 2340–2368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, C.C.C.R.; Fernandes, P. Production of metabolites as bacterial responses to the marine environment. Mar. Drugs 2010, 8, 705–727. [Google Scholar] [CrossRef] [PubMed]
- Lopanik, N.; Lindquist, N.; Targett, N. Potent cytotoxins produced by a microbial symbiont protect host larvae from predation. Oecologia 2004, 139, 131–139. [Google Scholar] [CrossRef]
- Kelman, D.; Kashman, Y.; Rosenberg, E.; Kushmaro, A.; Loya, Y. Antimicrobial activity of red sea corals. Mar. Biol. 2006, 149, 357–363. [Google Scholar] [CrossRef]
- Wei, W.C.; Sung, P.J.; Duh, C.Y.; Chen, B.W.; Sheu, J.H.; Yang, N.S. Anti-inflammatory activities of natural products Isolated from soft corals of Taiwan between 2008 and 2012. Mar. Drugs 2013, 11, 4083–4126. [Google Scholar] [CrossRef] [PubMed]
- Van de Water, J.A.J.M.; Allemand, D.; Ferrier-Pagès, C. Host-microbe interactions in octocoral holobionts—Recent advances and perspectives. Microbiome 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blunt, J.W.; Copp, B.R.; Hu, W.P.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2008, 25, 35–94. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Hu, W.P.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2009, 26, 170–244. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2014, 31, 160–258. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2013, 30, 237–323. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2012, 29, 144–222. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2011, 28, 196–268. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2010, 27, 165–237. [Google Scholar] [CrossRef] [PubMed]
- Bourne, D.G.; Morrow, K.M.; Webster, N.S. Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 2016, 70, 317–340. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Koren, O.; Reshef, L.; Efrony, R.; Zilber-Rosenberg, I. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 2007, 5, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, R.S.; Rosado, P.M.; Leite, D.C.A.; Rosado, A.S.; Bourne, D.G. Beneficial microorganisms for corals (BMC): Proposed mechanisms for coral health and resilience. Front. Microbiol. 2017, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.; Brugler, M.R.; Cartwright, P.; Collins, A.G.; Dawson, M.N.; Fautin, D.G.; France, S.C.; McFadden, C.S.; Opresko, D.M.; Rodriquez, E.; et al. The phylum Cnidaria: A review of phylogenetic patterns and diversity 300 years after Linnaeus. Zootaxa 2007, 1668, 127–182. [Google Scholar]
- Gili, J.M.; Garcia, A. Biologia de Paramuricea clavata a les costes catalanes generals. Bull. Inst. Cato Hist. Nat. 1985, 52, 25–32. [Google Scholar]
- Curdia, J.; Monteiro, P.; Afonso, C.M.L.; Santos, M.N.; Cunha, M.R.; Goncalves, J.M.S. Spatial and depth-associated distribution patterns of shallow gorgonians in the Algarve coast (Portugal, NE Atlantic). Helgol. Mar. Res. 2013, 67, 521–534. [Google Scholar] [CrossRef]
- Gili, J.M.; Coma, R. Benthic suspension feeders: Their paramount role in littoral marine food webs. Trends Ecol. Evolution. 1998, 13, 316–321. [Google Scholar] [CrossRef]
- Cerrano, C.; Danovaro, R.; Gambi, C.; Pusceddu, A.; Riva, A.; Schiaparelli, S. Gold coral (Savalia savaglia) and gorgonian forests enhance benthic biodiversity and ecosystem functioning in the mesophotic zone. Biodivers. Conserv. 2010, 19, 153–167. [Google Scholar] [CrossRef] [Green Version]
- Raina, J.B.; Tapiolas, D.; Willis, B.L.; Bourne, D.G. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 2009, 75, 3492–3501. [Google Scholar] [CrossRef] [PubMed]
- Ransome, E.; Rowley, S.J.; Thomas, S.; Tait, K.; Munn, C.B. Disturbance to conserved bacterial communities in the cold-water gorgonian coral Eunicella verrucosa. FEMS Microbiol. Ecol. 2014, 90, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Gori, A.; Bramanti, L.; Lopez-Gonzalez, P.; Thoma, J.N.; Gili, J.M.; Grinyo, J.; Uceira, V.; Rossi, S. Characterization of the zooxanthellate and azooxanthellate morphotypes of the Mediterranean gorgonian Eunicella singularis. Mar. Biol. 2012, 159, 1485–1496. [Google Scholar] [CrossRef]
- Keller-Costa, T.; Eriksson, D.; Gonçalves, J.M.S.; Gomes, N.C.M.; Lago-Leston, A.; Costa, R. The gorgonian coral Eunicella labiata hosts a distinct prokaryotic consortium amenable to cultivation. FEMS Microbiol. Ecol. 2017, 93, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bayer, T.; Arif, C.; Ferrier-Pages, C.; Zoccola, D.; Aranda, M.; Voolstra, C.R. Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini. Mar. Ecol. Prog. Ser. 2013, 479, 75–84. [Google Scholar] [CrossRef]
- La Riviere, M.; Roumagnac, M.; Garrabou, J.; Bally, M. Transient shifts in bacterial communities associated with the temperate gorgonian Paramuricea clavata in the Northwestern Mediterranean Sea. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Van de Water, J.A.J.M.; Melkonian, R.; Junca, H.; Voolstra, C.R.; Reynaud, S.; Allemand, D.; Ferrier-Pagès, C. Spirochaetes dominate the microbial community associated with the red coral Corallium rubrum on a broad geographic scale. Sci. Rep. 2016, 6, 27277. [Google Scholar] [CrossRef] [PubMed]
- Rath, C.; Janto, B.; Earl, J.; Ahmed, A.; Hu, F.Z.; Hiller, L.; Dahlgren, M.; Kreft, R.; Yu, F.; Wolff, J.J.; et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem. Biol. 2011, 6, 1244–1256. [Google Scholar] [CrossRef]
- Trindade, M.; van Zyl, L.J.; Navarro-Fernández, J.; Elrazak, A.A. Targeted metagenomics as a tool to tap into marine natural product diversity for the discovery and production of drug candidates. Front. Microbiol. 2015, 6, 890. [Google Scholar] [CrossRef]
- Barrero-Canosa, J.; Dueñas, L.F.; Sánchez, J.A. Isolation of potential fungal pathogens in gorgonian corals at the Tropical Eastern Pacific. Coral Reefs 2013, 32, 35–41. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Bao, J.; Wang, G.-H.; He, F.; Xu, X.-Y.; Qi, S.-H. Diversity and antimicrobial activity of culturable fungi isolated from six species of the South China Sea gorgonians. Microb. Ecol. 2012, 64, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Soler-Hurtado, M.M.; Sandoval-Sierra, J.V.; Annie Machordom, A.; DieÂguez-Uribeondo, J. Aspergillus sydowii and other potential fungal pathogens in gorgonian octocorals of the Ecuadorian Pacific. PLoS ONE 2016, 11, e0165992. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.C.; Shaw, H.; Rhodes, V.; Hart, A. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 2016, 7, 1728. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.; Smith, D.L.; Laxminarayan, R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 2007, 13, 1840–1846. [Google Scholar] [CrossRef] [PubMed]
- Boswihi, S.S.; Udo, E.E. Methicillin-resistant Staphylococcus aureus: An update on the epidemiology, treatment options and infection control. Curr. Med. Res. Pract. 2018, 8, 18–24. [Google Scholar] [CrossRef]
- Moree, W.J.; McConnell, O.J.; Nguyen, D.D.; Sanchez, L.M.; Yang, Y.-L.; Zhao, X.; Liu, W.-T.; Boudreau, P.D.; Srinivasan, J.; Atencio, L.; et al. Microbiota of healthy corals are active against fungi in a light-dependent manner. ACS Chem. Biol. 2014, 9, 2300–2308. [Google Scholar] [CrossRef] [PubMed]
- Shigemori, H.; Bae, M.-A.; Yazawa, K.; Sasaki, T.; Kobayashi, J. Alteramide A, a new tetracyclic alkaloid from a bacterium Alteromonas sp. associated with the marine sponge Ealhhondria okadai. J. Organ. Chem. 1992, 57, 4317–4320. [Google Scholar] [CrossRef]
- Gao, C.-H.; Tian, X.-P.; Qi, S.-H.; Luo, X.-M.; Wang, P.; Zhang, S. Antibacterial and antilarval compounds from marine gorgonian-associated bacterium Bacillus amyloliquefaciens SCSIO 00856. J. Antibiot. 2010, 63, 191–193. [Google Scholar] [CrossRef] [PubMed]
- Al-Zereini, W.; Yao, B.C.F.F.; Laatsch, H.; Anke, H. Aqabamycins A-G: Novel nitro maleimides from a marine Vibrio species: I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 2010, 63, 297–301. [Google Scholar] [CrossRef]
- Sulistiyani, S.; Nugraheni, S.A.; Radjasa, O.K.; Sabdono, A.; Khoeri, M.M. Antibacterial activities of bacterial symbionts of soft coral Sinularia sp. against tuberculosis bacteria. J. Coast. Dev. 2010, 14, 45–50. [Google Scholar]
- Radjasa, O.K.; Sabdono, A. Ecological role of a soft coral-associated bacterium Arthrobacter sp. on marine biofilm-forming bacteria. Microbiol. Indones. 2008, 2, 84–88. [Google Scholar]
- Zheng, C.J.; Shao, C.L.; Wu, L.Y.; Chen, M.; Wang, K.L.; Zhao, D.L.; Sun, X.P.; Chen, G.Y.; Wang, C.Y. Bioactive phenylalanine derivatives and cytochalasins from the soft coral-derived fungus, Aspergillus elegans. Mar. Drugs 2013, 11, 2054–2068. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.; Davies, A.P.; Harris, L.G.; Jeeves, R.; Pascoe, B.; Knobloch, J.K.M.; Rohde, H.; Wilkinson, T.S. Staphylococcus epidermidis in biomaterial-associated infections. In Biomaterials Associated Infection: Immunological Aspects and Antimicrobial Strategies; Moriarty, T.F., Zaat, S.A.J., Busscher, H.J., Eds.; Springer Science + Business Media: New York, NY, USA, 2013; pp. 25–56. [Google Scholar]
- Chen, M.; Fu, X.M.; Kong, C.J.; Wang, C.Y. Nucleoside derivatives from the marine-derived fungus Aspergillus versicolor. Nat. Prod. Res. 2014, 28, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Shao, C.L.; Fu, X.M.; Kong, C.J.; She, Z.G.; Wang, C.Y. Lumazine peptides penilumamides B-D and the cyclic pentapeptide asperpeptide A from a gorgonian-derived Aspergillus sp. fungus. J. Nat. Prod. 2014, 77, 1601–1606. [Google Scholar] [CrossRef]
- Wei, M.Y.; Wang, C.Y.; Liu, Q.A.; Shao, C.L.; She, Z.G.; Lin, Y.C. Five sesquiterpenoids from a marine-derived fungus Aspergillus sp. isolated from a gorgonian Dichotella gemmacea. Mar. Drugs 2010, 8, 941–949. [Google Scholar] [CrossRef]
- Khamthong, N.; Rukachaisirikul, V.; Phongpaichit, S.; Preedanon, S.; Sakayaroj, J. Bioactive polyketides from the sea fan-derived fungus Penicillium citrinum PSU-F51. Tetrahedron 2010, 68, 8245–8250. [Google Scholar] [CrossRef]
- Wang, J.; Liu, P.; Wang, Y.; Wang, H.; Li, J.; Zhuang, Y.; Zhu, W. Antimicrobial aromatic polyketides from gorgonian-associated fungus, Penicillium commune 518. Chin. J. Chem. 2012, 30, 1236–1242. [Google Scholar] [CrossRef]
- Wei, M.Y.; Li, D.; Shao, C.L.; Deng, D.S.; Wang, C.Y. (±)-Pestalachloride D, an antibacterial racemate of chlorinated benzophenone derivative from a soft coral-derived fungus Pestalotiopsis sp. Mar. Drugs 2013, 11, 1050–1060. [Google Scholar] [CrossRef]
- Zhuang, Y.; Teng, X.; Wang, Y.; Liu, P.; Wang, H.; Li, J.; Li, G.; Zhu, W. Cyclopeptides and polyketides from coral-associated fungus, Aspergillus versicolor LCJ-5-4. Tetrahedron 2011, 67, 7085–7089. [Google Scholar] [CrossRef]
- Zhuang, Y.; Teng, X.; Wang, Y.; Liu, P.; Li, G.; Zhu, W. New quinazolinone alkaloids within rare amino acid residue from coral- associated fungus, Aspergillus versicolor LCJ-5-4. Organ. Lett. 2011, 13, 1130–1133. [Google Scholar] [CrossRef]
- Satheesh, S.; Ba-akdah, M.A.; Al-Sofyani, A.A. Natural antifouling compound production by microbes associated with marine macroorganisms—A review. Electron. J. Biotechnol. 2016, 21, 26–35. [Google Scholar] [CrossRef]
- Antizar-Ladislao, B. Environmental levels, toxicity and human exposure to tributyltin (TBT)-contaminated marine environment. A review. Environ. Int. 2008, 34, 292–308. [Google Scholar] [CrossRef] [PubMed]
- Qian, P.-Y.; Li, Z.; Xu, Y.; Li, Y.; Fusetani, N. Mini-review: Marine natural products and their synthetic analogs as antifouling compounds: 2009–2014. Biofouling 2015, 31, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.-H.; Ma, X. Antifouling Compounds from Marine Invertebrates. Mar. Drugs 2017, 15, 263. [Google Scholar] [CrossRef]
- Dobretsov, S.; Qian, P.-Y. The role of epibotic bacteria from the surface of the soft coral Dendronephthya sp. in the inhibition of larval settlement. J. Exp. Mar. Biol. Ecol. 2004, 299, 35–50. [Google Scholar] [CrossRef]
- Shao, C.L.; Wu, H.X.; Wang, C.Y.; Liu, Q.A.; Xu, Y.; Wei, M.Y.; Qian, P.Y.; Gu, Y.C.; Zheng, C.J.; She, Z.G.; et al. Potent antifouling resorcylic acid lactones from the gorgonian-derived fungus Cochliobolus lunatus. J. Nat. Prod. 2011, 74, 629–633. [Google Scholar] [CrossRef]
- Bao, J.; Sun, Y.-L.; Zhang, X.-Y.; Han, Z.; Gao, H.-C.; He, F.; Qian, P.-Y.; Qi, S.-H. Antifouling and antibacterial polyketides from marine gorgonian coral-associated fungus Penicillium sp. SCSGAF 0023. J. Antibiot. 2012, 66, 219. [Google Scholar] [CrossRef]
- Bao, J.; Zhang, X.-Y.; Xu, X.-Y.; He, F.; Nong, X.-H.; Qi, S.-H. New cyclic tetrapeptides and asteltoxins from gorgonian-derived fungus Aspergillus sp. SCSGAF 0076. Tetrahedron 2013, 69, 2113–2117. [Google Scholar] [CrossRef]
- He, F.; Han, Z.; Peng, J.; Qian, P.-Y.; Qi, S.-H. Antifouling indole alkaloids from two marine derived fungi. Nat. Prod. Commun. 2013, 8, 329–332. [Google Scholar]
- Cheung, R.C.; Wong, J.H.; Pan, W.L.; Chan, Y.S.; Yin, C.M.; Dan, X.L.; Wang, H.X.; Fang, E.F.; Lam, S.K.; Ngai, P.H.; et al. Antifungal and antiviral products of marine organisms. Appl. Microbiol. Biotechnol. 2014, 98, 3475–3494. [Google Scholar] [CrossRef] [Green Version]
- Moghadamtousi, S.Z.; Nikzad, S.; Kadir, H.A.; Abubakar, S.; Keivan Zandi, K. Potential antiviral agents from marine fungi: An overview. Mar. Drugs 2015, 13, 4520–4538. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Shao, C.L.; Meng, H.; She, Z.G.; Wang, C.Y. Anti-respiratory syncytial virus prenylated dihydroquinolone derivatives from the gorgonian-derived fungus Aspergillus sp. XS-20090B15. J. Nat. Prod. 2014, 77, 2720–2724. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Bao, J.; Zhang, X.-Y.; Tu, Z.-C.; Shi, Y.-M.; Qi, S.-H. Asperterrestide A, a cytotoxic cyclic tetrapeptide from the marine-derived fungus Aspergillus terreus SCSGAF0162. J. Nat. Prod. 2013, 76, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Xu, X.-Y.; Zhang, X.-Y.; Qi, S.-H. A new macrolide from a marine-derived fungus Aspergillus sp. Nat. Prod. Commun. 2013, 8, 1127–1128. [Google Scholar] [PubMed]
- Nong, X.H.; Wang, Y.F.; Zhang, X.Y.; Zhou, M.P.; Xu, X.Y.; Qi, S.H. Territrem and butyrolactone derivatives from a marine-derived fungus Aspergillus terreus. Mar. Drugs 2014, 12, 6113–6124. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.-J.; Shao, C.-L.; Guo, Z.-Y.; Chen, J.-F.; Deng, D.-S.; Yang, K.-L.; Chen, Y.-Y.; Fu, X.-M.; She, Z.-G.; Lin, Y.-C.; et al. Bioactive hydroanthraquinones and anthraquinone dimers from a soft coral-derived Alternaria sp. fungus. J. Nat. Prod. 2012, 75, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.-L.; Wei, M.-Y.; Chen, H.-Y.; Guan, F.-F.; Wang, C.-Y.; Shao, C.-L. (+)- and (−)-Pestaloxazine A, a pair of antiviral enantiomeric alkaloid dimers with a symmetric spiro [oxazinane-piperazinedione] skeleton from Pestalotiopsis sp. Organ. Lett. 2015, 17, 4216–4219. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Torres, V.; Encinar, J.A.; Herranz-López, M.; Pérez-Sánchez, A.; Galiano, V.; Barrajón-Catalán, E.; Micol, V. An updated review on marine anticancer compounds: The use of virtual screening for the discovery of small-molecule cancer drugs. Molecules 2017, 22, 1037. [Google Scholar] [CrossRef]
- Grote, D.; Hänel, F.; Dahse, H.M.; Seifert, K. Capnellenes from the soft coral Dendronephthya rubeola. Chem. Biodivers. 2008, 5, 1683–1693. [Google Scholar] [CrossRef]
- Marrero, J.; Rodríguez, A.D.; Baran, P.; Raptis, R.G.; Sánchez, J.A.; Ortega-Barria, E.; Capson, T.L. Bielschowskysin, a gorgonian-derived biologically active diterpene with an unprecedented carbon skeleton. Organ. Lett. 2004, 6, 1661–1664. [Google Scholar] [CrossRef]
- Duh, C.Y.; El-Gamal, A.A.H.; Chu, C.J.; Wang, S.K.; Dai, C.F. New cytotoxic constituents from the Formosan soft corals Clavularia viridis and Clavularia violacea. J. Nat. Prod. 2002, 65, 1535–1539. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.C.; Cheng, Y.B.; Lin, Y.C.; Guh, J.H.; Teng, C.M.; Ko, C.L. New prostanoids with cytotoxic activity from Taiwanese octocoral Clavularia viridis. J. Nat. Prod. 2004, 67, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.-H.; Huang, L.-F.; Yang, Y.-L.; Su, J.-H.; Wang, G.-H.; Chiang, M.-Y.; Wu, Y.-C.; Dai, C.-F.; Sheu, J.H. Polyoxygenated steroids from the gorgonian Isis hippuris. J. Nat. Prod. 2005, 68, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.W.; Wu, Y.C.; Chiang, M.-Y.; Su, J.-H.; Wang, W.H.; Fan, T.-Y.; Sheu, J.H. Eunicellin-based diterpenoids from the cultured soft coral Klyxum simplex. Tetrahedron 2009, 65, 7016–7022. [Google Scholar] [CrossRef]
- Fu, P.; Kong, F.; Wang, Y.; Wang, Y.; Liu, P.; Zuo, G.; Zhu, W. Antibiotic metabolites from the coral-associated actinomycete Streptomyces sp. OUCMDZ-1703. Chin. J. Chem. 2013, 31, 100–104. [Google Scholar] [CrossRef]
- Chen, Y.-H.; Lu, M.-C.; Chang, Y.-C.; Hwang, T.-L.; Wang, W.H.; Weng, C.F.; Kuo, J.; Sung, P.J. Pseudoalteromone A: A novel bioactive ubiquinone from a marine bacterium Pseudoalteromonas sp. CGH2XX (Pseudoalteromonadaceae). Tetrahedron Lett. 2012, 53, 1675–1677. [Google Scholar] [CrossRef]
- Li, H.-J.; Jiang, W.-H.; Liang, W.-L.; Huang, J.-X.; Mo, Y.-F.; Ding, Y.-Q.; Lam, C.-K.; Qian, X.-J.; Zhu, X.-Z.; Lan, W.-J. Induced marine fungus Chondrostereum sp. as a means of producing new sesquiterpenoids chondrosterins I and J by using glycerol as the carbon source. Mar. Drugs 2014, 12, 167–175. [Google Scholar] [CrossRef]
- Li, H.-J.; Xie, Y.-L.; Xie, Z.-L.; Chen, Y.; Lam, C.-K.; Lan, W.-J. Chondrosterins A–E, triquinane-type sesquiterpenoids from soft coral-associated fungus Chondrostereum sp. Mar. Drugs 2012, 10, 627–638. [Google Scholar] [CrossRef]
- Li, H.-J.; Chen, T.; Xie, Y.-L.; Chen, W.-D.; Zhu, X.-F.; Lan, W.-J. Isolation and structural elucidation of chondrosterins F–H from the marine fungus Chondrostereum sp. Mar. Drugs 2013, 11, 551–558. [Google Scholar] [CrossRef]
- Li, H.-J.; Lan, W.-J.; Lam, C.-K.; Yang, F.; Zhu, X.-F. Hirsutane sesquiterpenoids from the marine-derived fungus Chondrostereum sp. Chem. Biodivers. 2011, 8, 317–324. [Google Scholar] [CrossRef]
- Shao, C.-L.; Wanga, C.-Y.; Wei, M.-Y.; Gu, Y.-C.; She, Z.-G.; Qian, P.-Y.; Lin, Y.-C. Aspergilones A and B, two benzylazaphilones with an unprecedented carbon skeleton from the gorgonian-derived fungus Aspergillus sp. Bioorgan. Med. Chem. Lett. 2011, 21, 690–693. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-L.; Bao, J.; Liu, K.-S.; Zhang, X.-Y.; He, F.; Wang, Y.F.; Nong, X.-H.; Qi, S.-H. Cytotoxic dihydrothiophene-condensed chromones from the marine-derived fungus Penicillium oxalicum. Planta Med. 2013, 79, 1474–1479. [Google Scholar] [CrossRef] [PubMed]
- Kasettrathat, C.; Ngamrojanavanich, N.; Wiyakrutta, S.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. Cytotoxic and antiplasmodial substances from marine-derived fungi, Nodulisporium sp. and CRI247-01. Phytochemistry 2008, 69, 2621–2626. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.S.; Glaser, K.B.; Cuevas, C.; Jacobs, R.S.; Kem, W.; Little, R.D.; McIntosh, J.M.; Newman, D.J.; Potts, B.C.; Shuster, D.E. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol. Sci. 2010, 31, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Mydlarz, L.D.; Jacobs, R.S.; Boehnlein, J.; Kerr, R.G. Pseudopterosin biosynthesis in Symbiodinium sp., the dinoflagellate symbiont of Pseudopterogorgia elisabethae. Chem. Biol. 2003, 10, 1051–1056. [Google Scholar] [CrossRef] [PubMed]
- Hofseth, L.J. Nitric oxide as a target of complementary and alternative medicines to prevent and treat inflammation and cancer. Cancer Lett. 2008, 268, 10–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Zhou, Q.; Wang, J.; Liu, J.; Qi, C.; Lai, Y.; Zhu, H.; Xue, Y.; Hu, Z.; Zhang, Y. Anti-inflammatory butenolide derivatives from the coral-derived fungus Aspergillus terreus and structure revisions of aspernolides D and G, butyrolactone VI and 4′,8′′-diacetoxy butyrolactone VI. RSC Adv. 2018, 8, 13040–13047. [Google Scholar] [CrossRef] [Green Version]
- Wei, M.-Y.; Chen, G.-Y.; Wang, Y.; Zhang, X.-L.; Wang, C.-Y.; Shao, C.-L. Isolation, 1H, 13C NMR assignments, and crystal structure of chrodrimanin B from a marine fungus Aspergillus sp. Chem. Nat. Compd. 2011, 47, 571–573. [Google Scholar] [CrossRef]
- Xu, Y.; Furutani, S.; Ihara, M.; Ling, Y.; Yang, X.; Kai, K.; Hayashi, H.; Matsuda, K. Meroterpenoid chrodrimanins are selective and potent blockers of insect GABA-gated chloride channels. PLoS ONE 2015, 10, e0122629. [Google Scholar] [CrossRef] [PubMed]
- Afiyatullov, S.S.; Zhuravleva, O.I.; Chaikina, E.L.; Anisimov, M.M. A new spirotryprostatin from the marine isolate of the fungus Aspergillus fumigatus. Chem. Nat. Compd. 2012, 48. [Google Scholar] [CrossRef]
- Martin, Y.; Bonnefort, J.L.; Chancerelle, L. Gorgonians mass mortality during the 1999 late summer in French Mediterranean coastal waters: The bacterial hypothesis. Water Res. 2002, 36, 779–782. [Google Scholar] [CrossRef]
- Hall-Spencer, J.M.; Pike, J.; Munn, C.B. Diseases affect cold-water corals too: Eunicella verrucosa (Cnidaria: Gorgonacea) necrosis in SW England. Dis. Aquatic Organ. 2007, 76, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Vezzulli, L.; Previati, M.; Pruzzo, C.; Marchese, A.; Bourne, D.G.; Cerrano, C.; VibrioSea, C. Vibrio infections triggering mass mortality events in a warming Mediterranean Sea. Environ. Microbiol. 2010, 12, 2007–2019. [Google Scholar] [CrossRef] [PubMed]
- Roussis, V.; Fenical, W.; Vagias, C.; Kornprobst, J.M.; Miralles, J. Labiatamides A, B, and other eunicellan diterpenoids from the Senegalese gorgonian Eunicella labiata. Tetrahedron 1996, 52, 2735–2742. [Google Scholar] [CrossRef]
- Keller-Costa, T.; Silva, R.; Lago-Lestón, A.; Costa, R. Genomic insights into Aquimarina sp. EL33, a bacterial symbiont of the gorgonian coral Eunicella labiata. Genome Announc. 2016, 4, e00855-16. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.G.; Lago-Lestón, A.; Costa, R.; Keller-Costa, T. Draft genome sequence of Sphingorhabdus sp. strain EL138, a metabolically-versatile alphaproteobacterium isolated from the gorgonian coral Eunicella labiata. Genome Announc. 2018, 6, e00142-18. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, G.N.; Lago-Lestón, A.; Costa, R.; Keller-Costa, T. Draft genome sequence of Labrenzia sp. strain EL143, a coral-associated alphaproteobacterium with versatile symbiotic living capability and strong halogen degradation potential. Genome Announc. 2018, 6, e00132-18. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Franco, T.; Califano, G.; Gonçalves, A.C.; Cúcio, C.; Costa, R. Draft genome sequence of Vibrio sp. strain Evh12, a bacterium retrieved from the gorgonian coral Eunicella verrucosa. Genome Announc. 2016, 4, e01729-15. [Google Scholar] [CrossRef] [PubMed]
- Cúcio, A.C. Molecular Exploration of Bacterial Communities Associated with Azooxanthellate Gorgonians in the Coast of Algarve, South Portugal; University of Algarve: Faro, Portugal, 2012. [Google Scholar]
- Ambati, R.R.; Phang, S.M.; Ravi, S.; Aswathanarayana, R.G. Astaxanthin: Sources, extraction, stability, biological activities and its commercial applications—A review. Mar. Drugs 2014, 12, 128. [Google Scholar] [CrossRef]
- Lucas-Elío, P.; Gómez, D.; Solano, F.; Sanchez-Amat, A. The antimicrobial activity of marinocine, synthesized by Marinomonas mediterranea, is due to hydrogen peroxide generated by its lysine oxidase activity. J. Bacteriol. 2006, 188, 2493–2501. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Peng, C.; Zhao, Y.; Li, Z. Functional gene-guided discovery of type II polyketides from culturable Actinomycetes associated with soft coral Scleronephthya sp. PLoS ONE 2012, 7, e42847. [Google Scholar] [CrossRef] [PubMed]
- Jakeman, D.L.; Bandi, S.; Graham, C.L.; Reid, T.R.; Wentzell, J.R.; Douglas, S.E. Antimicrobial activities of jadomycin B and structurally related analogues. Antimicrob. Agents Chemother. 2009, 53, 1245–1247. [Google Scholar] [CrossRef] [PubMed]
- Trincone, A. Marine biocatalysts: Enzymatic features and applications. Mar. Drugs 2011, 9, 478–499. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.; Kramer, A.; Labes, A.; Tasdemir, D. From discovery to production: Biotechnology of marine fungi for the production of new antibiotics. Mar. Drugs 2016, 14, 137. [Google Scholar] [CrossRef] [PubMed]
- Parente, R.; Feola, V.C.; Gimigliano, A. R&D management in the pharma industry: The strategic role of CROs. Sinerg. Ital. J. Manag. 2016, 34. [Google Scholar] [CrossRef]
- Pattnaik, P.; Pandey, S.C. University spinoffs: What, why, and how? Technol. Innov. Manag. Rev. 2014, 4, 44–50. [Google Scholar] [CrossRef]
- Park, S.Y.; Yang, D.; Ha, S.H.; Lee, S.Y. Metabolic engineering of microorganisms for the production of natural compounds. Adv. Biosyst. 2018, 2, 1700190. [Google Scholar] [CrossRef]
- Hjort, K.; Bergström, M.; Adesina, M.F.; Jansson, J.K.; Smalla, K.; Sjöling, S. Chitinase genes revealed and compared in bacterial isolates, DNA-extracts and a metagenomic library from a phytopathogen-suppressive soil. FEMS Microbiol. Ecol. 2010, 71, 197–207. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; Costa, R.; Jansson, J.; Sjöling, S.; Bailey, M.; Nalin, R.; Vogel, T.M.; van Overbeek, L. The metagenomics of disease-suppressive soils—Experiences from the METACONTROL project. Trends Biotechnol. 2008, 26, 591–601. [Google Scholar] [CrossRef]






| Category | Accession | Name | Description | Number of Open Reading Frames Detected per Gorgonian-Associated Bacterial Genome | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EL01 | EL26 | EL27 | EL44 | EL53 | EL129 | EL143 | EL138 | EL199 | EL58 | EL33 | Evd3 | Evd11 | Evh12 | Evh13 | ||||
| Monoterpene synthesis and metabolism | PF03088.9 | Str_synth | Strictosidine synthase | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| PF07858.5 | LEH | Limonene-1,2-epoxide hydrolase catalytic domain | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Tri- and tetraterpene synthesis | PF00494.12/COG1562 | SQS_PSY/ERG9 | Squalene/phytoene synthase | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| COG1233 | COG1233 | Phytoene dehydrogenase and related proteins | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | |
| PF08491.3 | SE | Squalene epoxidase | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | |
| PF05834.5 | Lycopene_cycl | Lycopene cyclase protein | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | |
| PF07143.4 | CrtC | Hydroxyneurosporene synthase | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 2 | 2 | 2 | |
| Polyketide synthases | PF08392.5 | FAE1_CUT1_RppA | FAE1/Type III polyketide synthase-like protein | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| PF00195.12 | Chal_sti_synt_N | Chalcone and stilbene synthases, N-terminal domain | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |
| PF02797.8 | Chal_sti_synt_C | Chalcone and stilbene synthases, C-terminal domain | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| COG2761 | FrnE | Predicted dithiol-disulfide isomerase involved in polyketide biosynthesis | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | |
| COG3321 | COG3321 | Polyketide synthase modules and related proteins | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 11 | 0 | 0 | 11 | 2 | 1 | 1 | 1 | |
| COG5285 | COG5285 | Protein involved in biosynthesis of mitomycin antibiotics/polyketide fumonisin | 2 | 1 | 2 | 1 | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Polyketide cyclases | PF03364.13 | Polyketide_cyc | Polyketide cyclase/dehydrase and lipid transport | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| PF10604.2 | Polyketide_cyc2 | Polyketide cyclase/dehydrase and lipid transport | 5 | 3 | 2 | 4 | 1 | 4 | 8 | 3 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | |
| PF07366.5 | SnoaL | SnoaL-like polyketide cyclase | 26 | 8 | 8 | 11 | 7 | 5 | 20 | 9 | 1 | 0 | 16 | 15 | 6 | 15 | 2 | |
| Nonribosomal peptides | PF08415.3 | NRPS | Nonribosomal peptide synthase | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Amino-glycoside antibiotics | PF02522.7 | Antibiotic_NAT | Aminoglycoside 3-N-acetyltransferase | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| PF03992.9 | ABM | Antibiotic biosynthesis monooxygenase | 11 | 9 | 5 | 2 | 2 | 4 | 11 | 9 | 6 | 2 | 5 | 6 | 6 | 6 | 6 | |
| Polycyclic peptide antibiotics | PF04737.6 | Lant_dehyd_N | Lantibiotic dehydratase, N-terminus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| PF04738.6 | Lant_dehyd_C | Lantibiotic dehydratase, C-terminus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raimundo, I.; Silva, S.G.; Costa, R.; Keller-Costa, T. Bioactive Secondary Metabolites from Octocoral-Associated Microbes—New Chances for Blue Growth. Mar. Drugs 2018, 16, 485. https://doi.org/10.3390/md16120485
Raimundo I, Silva SG, Costa R, Keller-Costa T. Bioactive Secondary Metabolites from Octocoral-Associated Microbes—New Chances for Blue Growth. Marine Drugs. 2018; 16(12):485. https://doi.org/10.3390/md16120485
Chicago/Turabian StyleRaimundo, Inês, Sandra G. Silva, Rodrigo Costa, and Tina Keller-Costa. 2018. "Bioactive Secondary Metabolites from Octocoral-Associated Microbes—New Chances for Blue Growth" Marine Drugs 16, no. 12: 485. https://doi.org/10.3390/md16120485
APA StyleRaimundo, I., Silva, S. G., Costa, R., & Keller-Costa, T. (2018). Bioactive Secondary Metabolites from Octocoral-Associated Microbes—New Chances for Blue Growth. Marine Drugs, 16(12), 485. https://doi.org/10.3390/md16120485