Marine Bacterial Polysaccharide EPS11 Inhibits Cancer Cell Growth via Blocking Cell Adhesion and Stimulating Anoikis
Abstract
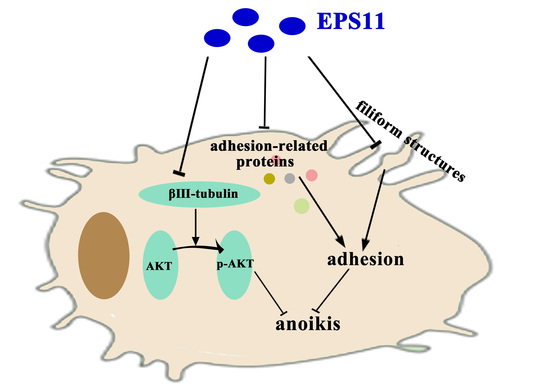
:1. Introduction
2. Results
2.1. Purification and Identification of Marine Bacterial Polysaccharide EPS11
2.2. EPS11 Preferentially Suppressed the Proliferation of Cancer Cells
2.3. EPS11 Suppressed Cell Adhesion in A549 Cells
2.4. EPS11 Destroyed Filiform Structures and Inhibited Cell Migration in A549 Cells
2.5. EPS11 Induced Apoptosis in A549 Cells
2.6. EPS11 Downregulated the Expression of βIII-Tubulin and Modulated AKT Activity
2.7. EPS11 Attenuated A549 Xenograft Tumor Growth In Vivo
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain Isolation, Identification, and Culture Conditions
4.2. Extraction, Isolation and Purification of Polysaccharides
4.3. Chemical Analysis of Cytotoxic Component of EPS11
4.4. Materials for Tissue Culture
4.5. Cell Proliferation Viability Assay
4.6. Cell Adhesion Viability Assay
4.7. Proteomic Analysis
4.8. Scanning Electron Microscope (SEM)
4.9. Transwell Migration Assay
4.10. Flow Cytometric Analysis of Apoptosis
4.11. Hoechst 33258 Staining
4.12. Quantitative Reverse Transcription-PCR (qRT-PCR)
4.13. Western Blot Analysis
4.14. Fluorescence Microscopy
4.15. Xenograft Tumor Model
4.16. Statistical Analysis
Acknowledgments
Author contributions
Conflicts of Interest
References
- Kumar, R.; Lu, S.K.; Minchom, A.; Sharp, A.; Davidson, M.; Gunapala, R.; Yap, T.A.; Bhosle, J.; Popat, S.; O’Brien, M.E.R. A phase 1b trial of the combination of an all-oral regimen of capecitabine and erlotinib in advanced non-small cell lung cancer in caucasian patients. Cancer Chemother. Pharm. 2016, 77, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Aisner, D.L.; Marshall, C.B. Molecular pathology of non–small cell lung cancer: A practical guide. Am. J. Clin. Pathol. 2012, 138, 332–346. [Google Scholar] [CrossRef] [PubMed]
- Goldstraw, P.; Ball, D.; Jett, J.R.; Le Chevalier, T.; Lim, E.; Nicholson, A.G.; Shepherd, F.A. Non-small-cell lung cancer. Lancet 2011, 378, 1727–1740. [Google Scholar] [CrossRef]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E. Nivolumab versus docetaxel in advanced nonsquamous non–small-cell lung cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.K.; Han, S.Q.; Xing, B.W.; Huang, J.Y.; Liu, B.Q.; Bordeleau, F.; Reinhart-King, C.A.; Zhang, J.J.; Huang, X.Y. Targeted inhibition of fascin function blocks tumour invasion and metastatic colonization. Nat. Commun. 2015, 6, 7468. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.D.; Anyiwe, K.; Schimmer, A.D. Anoikis resistance and tumor metastasis. Cancer Lett. 2008, 272, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, A.P. Anoikis. Cell Death Differ. 2005, 12, 1473–1477. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.A.; Paolillo, M.; Sanchez-Hernandez, Y.; Curti, D.; Ciusani, E.; Serra, M.; Colombo, L.; Schinelli, S. A small-molecule rgd-integrin antagonist inhibits cell adhesion, cell migration and induces anoikis in glioblastoma cells. Int. J. Oncol. 2013, 42, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Gumbiner, B.M. Cell adhesion: The molecular basis of tissue architecture and morphogenesis. Cell 1996, 84, 345–357. [Google Scholar] [CrossRef]
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Mogilner, A.; Rubinstein, B. The physics of filopodial protrusion. Biophys. J. 2005, 89, 782–795. [Google Scholar] [CrossRef] [PubMed]
- Mattila, P.K.; Lappalainen, P. Filopodia: Molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008, 9, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Sanders, T.A.; Llagostera, E.; Barna, M. Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature 2013, 497, 628. [Google Scholar] [CrossRef] [PubMed]
- Jacquemet, G.; Hamidi, H.; Ivaska, J. Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr. Opin. Cell Biol. 2015, 36, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wyckoff, J.B.; Frohlich, V.C.; Oleynikov, Y.; Huttelmaier, S.; Zavadil, J.; Cermak, L.; Bottinger, E.P.; Singer, R.H.; White, J.G.; et al. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling (vol 62, pg 6278, 2002). Cancer Res. 2002, 62, 7132. [Google Scholar]
- Berezovskaya, O.; Schimmer, A.D.; Glinskii, A.B.; Pinilla, C.; Hoffman, R.M.; Reed, J.C.; Glinsky, G.V. Increased expression of apoptosis inhibitor protein xiap contributes to anoikis resistance of circulating human prostate cancer metastasis precursor cells. Cancer Res. 2005, 65, 2378–2386. [Google Scholar] [CrossRef] [PubMed]
- McCarroll, J.A.; Gan, P.P.; Erlich, R.B.; Liu, M.; Dwarte, T.; Sagnella, S.S.; Akerfeldt, M.C.; Yang, L.; Parker, A.L.; Chang, M.H. Tubb3/βiii-tubulin acts through the pten/akt signaling axis to promote tumorigenesis and anoikis resistance in non–small cell lung cancer. Cancer Res. 2015, 75, 415–425. [Google Scholar] [CrossRef] [PubMed]
- McCarroll, J.A.; Sharbeen, G.; Liu, J.; Youkhana, J.; Goldstein, D.; McCarthy, N.; Limbri, L.F.; Dischl, D.; Ceyhan, G.O.; Erkan, M.; et al. βIII-tubulin: A novel mediator of chemoresistance and metastases in pancreatic cancer. Oncotarget 2015, 6, 2235–2249. [Google Scholar] [CrossRef] [PubMed]
- Gan, P.P.; Pasquier, E.; Kavallaris, M. Class iii β-tubulin mediates sensitivity to chemotherapeutic drugs in non–small cell lung cancer. Cancer Res. 2007, 67, 9356–9363. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, D.J.; Chen, J.; Wang, S.G. A polysaccharide from pinellia ternata inhibits cell proliferation and metastasis in human cholangiocarcinoma cells by targeting of cdc42 and 67 kda laminin receptor (lr). Int. J. Biol. Macromol. 2016, 93, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Varghese, S.; Joseph, M.M.; Aravind, S.R.; Unnikrishnan, B.S.; Sreelekha, T.T. The inhibitory effect of anti- tumor polysaccharide from punica granatum on metastasis. Int. J. Biol. Macromol. 2017, 103, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
- Ruocco, N.; Costantini, S.; Guariniello, S.; Costantini, M. Polysaccharides from the marine environment with pharmacological, cosmeceutical and nutraceutical potential. Molecules 2016, 21, 551. [Google Scholar] [CrossRef] [PubMed]
- Campbell, I.G.; Foulkes, W.D.; Senger, G.; Trowsdale, J.; Garinchesa, P.; Rettig, W.J. Molecular-cloning of the b-cam cell-surface glycoprotein of epithelial cancers—A novel member of the immunoglobulin superfamily. Cancer Res. 1994, 54, 5761–5765. [Google Scholar] [PubMed]
- Schiller, H.B.; Hermann, M.R.; Polleux, J.; Vignaud, T.; Zanivan, S.; Friedel, C.C.; Sun, Z.Q.; Raducanu, A.; Gottschalk, K.E.; Thery, M.; et al. β(1)- and α(v)-class integrins cooperate to regulate myosin ii during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 2013, 15, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.; Strugnell, S.S.; Goetz, J.G.; Kojic, L.D.; Cox, M.E.; Griffith, O.L.; Chan, S.K.; Jones, S.J.; Leung, S.P.; Masoudi, H.; et al. Phosphorylated caveolin-1 regulates rho/rock-dependent focal adhesion dynamics and tumor cell migration and invasion. Cancer Res. 2008, 68, 8210–8220. [Google Scholar] [CrossRef] [PubMed]
- Ikeo, K.; Oshima, T.; Shan, J.; Matsui, H.; Tomita, T.; Fukui, H.; Watari, J.; Miwa, H. Junctional adhesion molecule-a promotes proliferation and inhibits apoptosis of gastric cancer. Hepato-Gastroenterol. 2015, 62, 540–545. [Google Scholar]
- Fischer, R.S.; Yarmola, E.G.; Weber, K.L.; Speicher, K.D.; Speicher, D.W.; Bubb, M.R.; Fowler, V.M. Tropomodulin 3 binds to actin monomers. J. Biol. Chem. 2006, 281, 36454–36465. [Google Scholar] [CrossRef] [PubMed]
- Aktary, Z.; Alaee, M.; Pasdar, M. Beyond cell-cell adhesion: Plakoglobin and the regulation of tumorigenesis and metastasis. Oncotarget 2017, 8, 32270–32291. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Sheng, S.; Pardee, A.B. Metastasis and akt activation. Cell Cycle 2008, 7, 2991–2996. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, S.; Benetatos, C.A.; Colarusso, P.J.; Dexter, D.W.; Hudes, G.R. Altered beta-tubulin isotype expression in paclitaxel-resistant human prostate carcinoma cells. Br. J. Cancer 1998, 77, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Heymann, D.; Ruiz-Velasco, C.; Chesneau, J.; Ratiskol, J.; Sinquin, C.; Colliec-Jouault, S. Anti-metastatic properties of a marine bacterial exopolysaccharide-based derivative designed to mimic glycosaminoglycans. Molecules 2016, 21, 309. [Google Scholar] [CrossRef] [PubMed]
- Kavallaris, M.; Kuo, D.Y.-S.; Burkhart, C.A.; Regl, D.L.; Norris, M.D.; Haber, M.; Horwitz, S.B. Taxol-resistant epithelial ovarian tumors are associated with altered expression of specific beta-tubulin isotypes. J. Clin. Investig. 1997, 100, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Chanvorachote, P.; Pongrakhananon, V.; Halim, H. Caveolin-1 regulates metastatic behaviors of anoikis resistant lung cancer cells. Mol. Cell. Biochem. 2015, 399, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Weaver, A.M. Invadopodia: Specialized cell structures for cancer invasion. Clin. Exp. Metastasis 2006, 23, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Brisson, L.; Driffort, V.; Benoist, L.; Poet, M.; Counillon, L.; Antelmi, E.; Rubino, R.; Besson, P.; Labbal, F.; Chevalier, S. Nav1. 5 Na+ channels allosterically regulate the nhe-1 exchanger and promote the activity of breast cancer cell invadopodia. J. Cell Sci. 2013, 126, 4835–4842. [Google Scholar] [CrossRef] [PubMed]
- Tan, V.Y.; Lewis, S.J.; Adams, J.C.; Martin, R.M. Association of fascin-1 with mortality, disease progression and metastasis in carcinomas: A systematic review and meta-analysis. BMC Med. 2013, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Frisch, S.M.; Ruoslahti, E. Integrins and anoikis. Curr. Opin. Cell Biol. 1997, 9, 701–706. [Google Scholar] [CrossRef]
- Mozzetti, S.; Ferlini, C.; Concolino, P.; Filippetti, F.; Raspaglio, G.; Prislei, S.; Gallo, D.; Martinelli, E.; Ranelletti, F.O.; Ferrandina, G.; et al. Class III beta-tubulin overexpression is a prominent mechanism of paclitaxel resistance in ovarian cancer patients. Clin. Cancer Res. 2005, 11, 298–305. [Google Scholar] [PubMed]
- Egevad, L.; Valdman, A.; Wiklund, N.P.; Sève, P.; Dumontet, C. β-tubulin iii expression in prostate cancer. Scand. J. Urol. Nephrol. 2010, 44, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Seve, P.; Isaac, S.; Tredan, O.; Souquet, P.J.; Pacheco, Y.; Perol, M.; Lafanechere, L.; Penet, A.; Peiller, E.L.; Dumontet, C. Expression of class iii beta-tubulin is predictive of patient outcome in patients with non-small cell lung cancer receiving vinorelbine-based chemotherapy. Clin. Cancer Res. 2005, 11, 5481–5486. [Google Scholar] [CrossRef] [PubMed]
- Tommasi, S.; Mangia, A.; Lacalamita, R.; Bellizzi, A.; Fedele, V.; Chiriatti, A.; Thomssen, C.; Kendzierski, N.; Latorre, A.; Lorusso, V.; et al. Cytoskeleton and paclitaxel sensitivity in breast cancer: The role of beta-tubulins. Int. J. Cancer 2007, 120, 2078–2085. [Google Scholar] [CrossRef] [PubMed]
- Schut, F.; Devries, E.J.; Gottschal, J.C.; Robertson, B.R.; Harder, W.; Prins, R.A.; Button, D.K. Isolation of typical marine-bacteria by dilution culture—Growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microb. 1993, 59, 2150–2160. [Google Scholar]
- Xiu, P.Y.; Liu, R.; Zhang, D.C.; Sun, C.M. Pumilacidin-like lipopeptides derived from marineb bacterium bacillus sp. Strain 176 suppress the motility of Vibrio alginolyticus. Appl. Environ. Microb. 2017, 83, e00450-17. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, L.; Yao, J.; Shi, Y.; Li, P.; Ding, K. An arabinogalactan from flowers of Panax notoginseng inhibits angiogenesis by BMP2/Smad/Id1 signaling. Carbohydr. Polym. 2015, 121, 328–335. [Google Scholar] [CrossRef] [PubMed]










© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, R.; Jin, W.; Shan, Y.; Wang, J.; Liu, G.; Kuang, S.; Sun, C. Marine Bacterial Polysaccharide EPS11 Inhibits Cancer Cell Growth via Blocking Cell Adhesion and Stimulating Anoikis. Mar. Drugs 2018, 16, 85. https://doi.org/10.3390/md16030085
Cao R, Jin W, Shan Y, Wang J, Liu G, Kuang S, Sun C. Marine Bacterial Polysaccharide EPS11 Inhibits Cancer Cell Growth via Blocking Cell Adhesion and Stimulating Anoikis. Marine Drugs. 2018; 16(3):85. https://doi.org/10.3390/md16030085
Chicago/Turabian StyleCao, Ruobing, Weihua Jin, Yeqi Shan, Ju Wang, Ge Liu, Shan Kuang, and Chaomin Sun. 2018. "Marine Bacterial Polysaccharide EPS11 Inhibits Cancer Cell Growth via Blocking Cell Adhesion and Stimulating Anoikis" Marine Drugs 16, no. 3: 85. https://doi.org/10.3390/md16030085
APA StyleCao, R., Jin, W., Shan, Y., Wang, J., Liu, G., Kuang, S., & Sun, C. (2018). Marine Bacterial Polysaccharide EPS11 Inhibits Cancer Cell Growth via Blocking Cell Adhesion and Stimulating Anoikis. Marine Drugs, 16(3), 85. https://doi.org/10.3390/md16030085