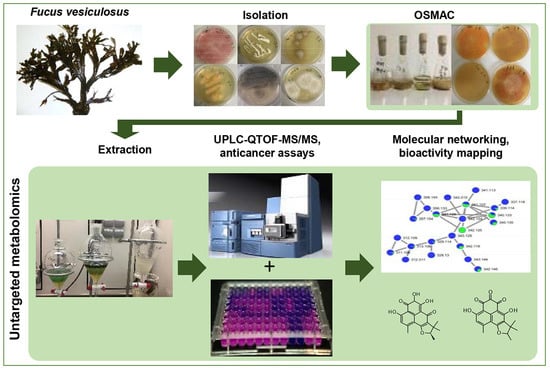
Influence of OSMAC-Based Cultivation in Metabolome and Anticancer Activity of Fungi Associated with the Brown Alga Fucus vesiculosus
Abstract
:1. Introduction
2. Results
2.1. Isolation of Fungi
2.2. OSMAC Cultivation, Extraction and Anticancer Assays
2.3. Evaluation of the Chemical Diversity Using Molecular Network Approach
2.3.1. Impact of Culture Regime on Chemical Diversity
2.3.2. Impact of Culture Media on Chemical Diversity
2.3.3. Bioactivity Mapping
3. Discussion
3.1. Fungal Isolation
3.2. Evaluation of Chemical Diversity Under Different Culture Conditions
3.3. Bioactivity
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Sampling and Isolation of Fungi
4.3. Identification of Fungal Strains
4.4. OSMAC-Based Cultivation of Fungi
4.5. Extraction
4.6. Chemical Analysis
4.6.1. UPLC-QToF-MS/MS Analysis
4.6.2. Molecular Network and Data Analysis
4.7. Anticancer Activity
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zuccaro, A.; Schoch, C.L.; Spatafora, J.W.; Kohlmeyer, J.; Draeger, S.; Mitchell, J.I. Detection and identification of fungi intimately associated with the brown seaweed Fucus serratus. Appl. Environ. Microbiol. 2008, 74, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Loque, C.P.; Medeiros, A.O.; Pellizzari, F.M.; Oliveira, E.C.; Rosa, C.A.; Rosa, L.H. Fungal community associated with marine macroalgae from Antarctica. Polar Biol. 2010, 33, 641–648. [Google Scholar] [CrossRef]
- Harvey, J.B.J.; Goff, L.J. Genetic covariation of the marine fungal symbiont Haloguignardia irritans (Ascomycota, Pezizomycotina) with its algal hosts Cystoseira and Halidrys (Phaeophyceae, Fucales) along the west coast of North America. Fungal Biol. 2010, 114, 82–95. [Google Scholar] [CrossRef] [PubMed]
- Egan, S.; Harder, T.; Burke, C.; Steinberg, P.; Kjelleberg, S.; Thomas, T. The seaweed holobiont: Understanding seaweed–bacteria interactions. FEMS Microbiol. Rev. 2013, 37, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Sarasan, M.; Puthumana, J.; Job, N.; Han, J.; Lee, J.-S.; Philip, R. Marine algicolous endophytic fungi-a promising drug resource of the era. J. Microbiol. Biotechnol. 2017, 27, 1039–1052. [Google Scholar] [PubMed]
- Flewelling, A.J.; Johnson, J.A.; Gray, C.A. Isolation and bioassay screening of fungal endophytes from North Atlantic marine macroalgae. Bot. Mar. 2013, 56, 287–297. [Google Scholar] [CrossRef]
- Flewelling, A.J.; Ellsworth, K.T.; Sanford, J.; Forward, E.; Johnson, J.A.; Gray, C.A. Macroalgal endophytes from the Atlantic coast of Canada: A potential source of antibiotic natural products? Microorganisms 2013, 1, 175–187. [Google Scholar] [CrossRef]
- Fenical, W.; Jensen, P.R.; Cheng, X.C. Halimide, a Cytotoxic Marine Natural Product, and Derivatives Thereof. U.S. Patent US6358957B1, 30 May 2000. [Google Scholar]
- Gomes, N.G.; Lefranc, F.; Kijjoa, A.; Kiss, R. Can some marine-derived fungal metabolites become actual anticancer agents? Mar. Drugs 2015, 13, 3950–3991. [Google Scholar] [CrossRef]
- Romano, S.; Jackson, S.; Patry, S.; Dobson, A. Extending the “One Strain Many Compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef]
- Reen, F.J.; Romano, S.; Dobson, A.D.; O’Gara, F. The sound of silence: Activating silent biosynthetic gene clusters in marine microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Gulder, T.; Hong, H.; Correa, J.; Egereva, E.; Wiese, J.; Imhoff, J.; Gross, H. Isolation, structure elucidation and total synthesis of lajollamide A from the marine fungus Asteromyces cruciatus. Mar. Drugs 2012, 10, 2912–2935. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; De Felicio, R.; Fenner, A. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [PubMed]
- Allard, P.-M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.-L. Integration of molecular networking and in-silico MS/MS fragmentation for natural products dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef] [PubMed]
- Crüsemann, M.; O’Neill, E.C.; Larson, C.B.; Melnik, A.V.; Floros, D.J.; da Silva, R.R.; Jensen, P.R.; Dorrestein, P.C.; Moore, B.S. Prioritizing natural product diversity in a collection of 146 bacterial strains based on growth and extraction protocols. J. Nat. Prod. 2016, 80, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Olivon, F.; Allard, P.-M.; Koval, A.; Righi, D.; Genta-Jouve, G.; Neyts, J.; Apel, C.; Pannecouque, C.; Nothias, L.-F.; Cachet, X. Bioactive natural products prioritization using massive multi-informational molecular networks. ACS Chem. Biol. 2017, 12, 2644–2651. [Google Scholar] [CrossRef] [PubMed]
- Ardehed, A.; Johansson, D.; Sundqvist, L.; Schagerström, E.; Zagrodzka, Z.; Kovaltchouk, N.A.; Bergström, L.; Kautsky, L.; Rafajlovic, M.; Pereyra, R.T.; et al. Divergence within and among seaweed siblings (Fucus vesiculosus and F. radicans) in the Baltic Sea. PLoS ONE 2016, 11, e0161266–e0161281. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L. A review of the nutrient composition of selected edible seaweeds. In Seaweed: Ecology, Nutrient Composition and Medicinal Uses; Nova Science: Coimbra, Portugal, 2011; pp. 15–47. [Google Scholar]
- Zenthoefer, M.; Geisen, U.; Hofmann-Peiker, K.; Fuhrmann, M.; Kerber, J.; Kirchhöfer, R.; Hennig, S.; Peipp, M.; Geyer, R.; Piker, L. Isolation of polyphenols with anticancer activity from the Baltic Sea brown seaweed Fucus vesiculosus using bioassay-guided fractionation. J. Appl. Phycol. 2017, 29, 2021–2037. [Google Scholar] [CrossRef]
- Heavisides, E.; Rouger, C.; Reichel, A.; Ulrich, C.; Wenzel-Storjohann, A.; Sebens, S.; Tasdemir, D. Seasonal variations in the metabolome and bioactivity profile of Fucus vesiculosus extracted by an optimised, pressurised liquid extraction protocol. Mar. Drugs 2018, 16, 503. [Google Scholar] [CrossRef]
- Abdel-Lateff, A.; Fisch, K.M.; Wright, A.D.; König, G.M. A new antioxidant isobenzofuranone derivative from the algicolous marine fungus Epicoccum sp. Planta Med. 2003, 69, 831–834. [Google Scholar] [PubMed]
- Whittle, M.; Willett, P.; Klaffke, W.; van Noort, P. Evaluation of similarity measures for searching the dictionary of natural products database. J. Chem. Inf. Comput. Sci. 2003, 43, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Ridley, D.D. Introduction to structure searching with SciFinder Scholar. J. Chem. Educ. 2001, 78, 559–560. [Google Scholar] [CrossRef]
- Little, J.L.; Williams, A.J.; Pshenichnov, A.; Tkachenko, V. Identification of “Known unknowns” utilizing accurate mass data and ChemSpider. J. Am. Soc. Mass Spectrom. 2012, 23, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Zuccaro, A.; Schulz, B.; Mitchell, J.I. Molecular detection of ascomycetes associated with Fucus serratus. Mycol. Res. 2004, 107, 1451–1466. [Google Scholar] [CrossRef]
- Godinho, V.M.; Furbino, L.E.; Santiago, I.F.; Pellizzari, F.M.; Yokoya, N.S.; Pupo, D.; Alves, T.M.A.; Junior, P.A.S.; Romanha, A.J.; Zani, C.L.; et al. Diversity and bioprospecting of fungal communities associated with endemic and cold-adapted macroalgae in Antarctica. ISME J. 2013, 7, 1434–1451. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Gawad, K.M.; Hifney, A.F.; Issa, A.A.; Gomaa, M. Spatio-temporal, environmental factors, and host identity shape culturable-epibiotic fungi of seaweeds in the Red Sea, Egypt. Hydrobiologia 2014, 740, 37–49. [Google Scholar] [CrossRef]
- Almeida, C.; Eguereva, E.; Kehraus, S.; Siering, C.; König, G.M. Hydroxylated sclerosporin derivatives from the marine-derived fungus Cadophora malorum. J. Nat. Prod. 2010, 73, 476–478. [Google Scholar] [CrossRef] [PubMed]
- Zalar, P.; De Hoog, G.S.; Schroers, H.-J.; Frank, J.M.; Gunde-Cimerman, N. Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Anton Leeuw 2005, 87, 311–328. [Google Scholar] [CrossRef]
- Cornax, R.; Moriñigo, M.A.; Romero, P.; Borrego, J.J. Survival of pathogenic microorganisms in seawater. Curr. Microbiol. 1990, 20, 293–298. [Google Scholar] [CrossRef]
- Nagahama, T.; Takahashi, E.; Nagano, Y.; Abdel-Wahab, M.A.; Miyazaki, M. Molecular evidence that deep-branching fungi are major fungal components in deep-sea methane cold-seep sediments. Environ. Microbiol. 2011, 13, 2359–2370. [Google Scholar] [CrossRef] [PubMed]
- Ţura, D.; Zmitrovich, I.V.; Wasser, S.P.; Nevo, E. The genus Stereum in Israel. Mycotaxon 2008, 106, 109–126. [Google Scholar]
- Rickert, E.; Lenz, M.; Barboza, F.R.; Gorb, S.N.; Wahl, M. Seasonally fluctuating chemical microfouling control in Fucus vesiculosus and Fucus serratus from the Baltic Sea. Mar. Biol. 2016, 163, 203–215. [Google Scholar] [CrossRef]
- Weinberger, F. Pathogen-induced defense and innate immunity in macroalgae. Biol. Bull. 2007, 213, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Sivagnanam, S.; Yin, S.; Choi, J.; Park, Y.; Woo, H.; Chun, B. Biological properties of fucoxanthin in oil recovered from two brown seaweeds using supercritical CO2 extraction. Mar. Drugs 2015, 13, 3422–3442. [Google Scholar] [CrossRef] [PubMed]
- Lachnit, T.; Fischer, M.; Künzel, S.; Baines, J.F.; Harder, T. Compounds associated with algal surfaces mediate epiphytic colonization of the marine macroalga Fucus vesiculosus. FEMS Microbiol. Ecol. 2013, 84, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Hewage, R.T.; Aree, T.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. One strain-many compounds (OSMAC) method for production of polyketides, azaphilones, and an isochromanone using the endophytic fungus Dothideomycete sp. Phytochemistry 2014, 108, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Papagianni, M. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 2004, 22, 189–259. [Google Scholar] [CrossRef] [PubMed]
- Bigelis, R.; He, H.; Yang, H.Y.; Chang, L.-P.; Greenstein, M. Production of fungal antibiotics using polymeric solid supports in solid-state and liquid fermentation. J. Ind. Microbiol. Biotechnol. 2006, 33, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Nout, M.J.R.; Bonants-van Laarhoven, T.M.G.; de Jongh, P.; de Koster, P.G. Ergosterol content of Rhizopus oligosporus NRRL 5905 grown in liquid and solid substrates. Appl. Microbiol. Biotechnol. 1987, 26, 456–461. [Google Scholar] [CrossRef]
- Guo, W.; Peng, J.; Zhu, T.; Gu, Q.; Keyzers, R.A.; Li, D. Sorbicillamines A–E, nitrogen-containing sorbicillinoids from the deep-sea-derived fungus Penicillium sp. F23–2. J. Nat. Prod. 2013, 76, 2106–2112. [Google Scholar] [CrossRef] [PubMed]
- Davis, N.D.; Diener, U.L.; Eldridge, D.W. Production of aflatoxins B1 and G1 by Aspergillus flavus in a semisynthetic medium. Appl. Microbiol. 1966, 14, 378–380. [Google Scholar] [PubMed]
- Rodríguez-Ortiz, R.; Mehta, B.J.; Avalos, J.; Limón, M.C. Stimulation of bikaverin production by sucrose and by salt starvation in Fusarium fujikuroi. Appl. Microbiol. Biotechnol. 2010, 85, 1991–2000. [Google Scholar] [CrossRef] [PubMed]
- Chatzifragkou, A.; Fakas, S.; Galiotou-Panayotou, M.; Komaitis, M.; Aggelis, G.; Papanikolaou, S. Commercial sugars as substrates for lipid accumulation in Cunninghamella echinulata and Mortierella isabellina fungi. Eur. J. Lipid Sci. Technol. 2010, 112, 1048–1057. [Google Scholar] [CrossRef]
- Måhlén, A. Purification and some properties of ATP citrate lyase from Penicillium spiculisporum. Eur. J. Biochem. 1973, 36, 342–346. [Google Scholar] [CrossRef]
- Muhid, F.; Nawi, W.; Kader, A.J.A.; Yusoff, W.M.W.; Hamid, A.A. Effects of metal ion concentrations on lipid and gamma linolenic acid production by Cunninghamella sp. 2A1. Online J. Biol. Sci. 2008, 8, 62–67. [Google Scholar] [CrossRef]
- Yang, X.; Kang, M.-C.; Li, Y.; Kim, E.-A.; Kang, S.-M.; Jeon, Y.-J. Anti-inflammatory activity of questinol isolated from marine-derived fungus Eurotium amstelodami in lipopolysaccharide-stimulated RAW 264.7 macrophages. J. Microbiol. Biotechnol 2014, 24, 1346–1353. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, S.; Marcos, A.T.; Casqueiro, J.; Kosalková, K.; Fernández, F.J.; Velasco, J.; Martín, J.F. Transcription of the pcbAB, pcbC and penDE genes of Penicillium chrysogenum AS-P-78 is repressed by glucose and the repression is not reversed by alkaline pHs. Microbiology 1999, 145, 317–324. [Google Scholar] [CrossRef]
- Wagener, R.E.; Davis, N.D.; Diener, U.L. Penitrem A and roquefortine production by Penicillium commune. Appl. Environ. Microbiol. 1980, 39, 882–887. [Google Scholar]
- Nagashima, H. Cytotoxic effects of rubratoxin B on cultured cells. JSM Myctoxins 1996, 1996, 57–61. [Google Scholar] [CrossRef]
- Moss, M.O.; Wood, A.B.; Robinson, F.V. The structure of rubratoxin A, a toxic metabolite of Penicillium rubrum. Tetrahedron Lett. 1969, 10, 367–370. [Google Scholar] [CrossRef]
- Wada, S.-I.; Usami, I.; Umezawa, Y.; Inoue, H.; Ohba, S.-I.; Someno, T.; Kawada, M.; Ikeda, D. Rubratoxin A specifically and potently inhibits protein phosphatase 2A and suppresses cancer metastasis. Cancer Sci. 2010, 101, 743–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanova, L.; Skjerve, E.; Eriksen, G.S.; Uhlig, S. Cytotoxicity of enniatins A, A1, B, B1, B2 and B3 from Fusarium avenaceum. Toxicon 2006, 47, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.-F.; Nothias-Esposito, M.; da Silva, R.; Wang, M.; Protsyuk, I.; Zhang, Z.; Sarvepalli, A.; Leyssen, P.; Touboul, D.; Costa, J.; et al. Bioactivity-based molecular networking for the discovery of drug leads in natural product bioassay-guided fractionation. J. Nat. Prod. 2018, 81, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Mefteh, F.B.; Daoud, A.; Bouket, A.C.; Thissera, B.; Kadri, Y.; Cherif-Silini, H.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Oszako, T. Date palm trees root-derived endophytes as fungal cell factories for diverse bioactive metabolites. Int. J. Mol. Sci. 2018, 19, 1986. [Google Scholar] [CrossRef]
- Wu, H.-Y.; Yang, F.-L.; Li, L.-H.; Rao, Y.K.; Ju, T.-C.; Wong, W.-T.; Hsieh, C.-Y.; Pivkin, M.V.; Hua, K.-F.; Wu, S.-H. Ergosterol peroxide from marine fungus Phoma sp. induces ROS-dependent apoptosis and autophagy in human lung adenocarcinoma cells. Sci. Rep. 2018, 8, 17956–17969. [Google Scholar] [CrossRef]
- Kobayashi, H.; Meguro, S.; Yoshimoto, T.; Namikoshi, M. Absolute structure, biosynthesis, and anti-microtubule activity of phomopsidin, isolated from a marine-derived fungus Phomopsis sp. Tetrahedron 2003, 59, 455–459. [Google Scholar] [CrossRef]
- Silber, J.; Ohlendorf, B.; Labes, A.; Erhard, A.; Imhoff, J.F. Calcarides A–E, antibacterial macrocyclic and linear polyesters from a Calcarisporium strain. Mar. Drugs 2013, 11, 3309–3323. [Google Scholar] [CrossRef]
- Atlas, R.M. Handbook of Microbiological Media, 4th ed.; CRC Press: Boca Raton, FL, USA, 2010; pp. 1654–1657. [Google Scholar]
- Schulz, B.; Wanke, U.; Draeger, S.; Aust, H.J. Endophytes from herbaceous plants and shrubs: Effectiveness of surface sterilization methods. Mycol. Res. 1993, 97, 1447–1450. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Wiese, J.; Ohlendorf, B.; Blümel, M.; Schmaljohann, R.; Imhoff, J.F. Phylogenetic identification of fungi isolated from the marine sponge Tethya aurantium and identification of their secondary metabolites. Mar. Drugs 2011, 9, 561–585. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ř.; Harper, D.; Ryan, P. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodraska, D.; Sanford, M.; Xu, D. Security mutation testing of the FileZilla FTP server. In Proceedings of the 2011 ACM Symposium on Applied Computing, TaiChung, Taiwan, 21–24 March 2011; pp. 1425–1430. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]








| Strain I.D. | Epiphytic/Endophytic Fungus | Closest Taxonomical Relative | Sequence Similarity (%) | Isolation Solid Medium |
|---|---|---|---|---|
| 1 * | Endophytic | Pleosporales (order) | 100 | WM-S |
| 11 | Epiphytic | Gibellulopsis sp. | 100 | GCM-S |
| 12 | Endophytic | Acremonium sp. | 99 | WM-S |
| 35 | Endophytic | Cadophora malorum | 100 | GYP-S |
| 37 * | Endophytic | Hypocreales (order) | 99 | GYP-S |
| 38 | Endophytic | Penicillium biourgeianum | 100 | WM-S |
| 50 | Endophytic | Penicillium sp. | 99 | PDM-S |
| 55 | Endophytic | Penicillium glabrum | 100 | PDM-S |
| 56 | Endophytic | Penicillium sp. | 100 | WM-S |
| 58 | Endophytic | Fusarium graminearum | 96 | WM-S |
| 59 * | Endophytic | Glomerellales (order) | 100 | WM-S |
| 61 | Endophytic | Penicillium brevicompactum | 100 | PDM-S |
| 62 | Endophytic | Penicillium brevicompactum | 100 | PDM-S |
| 67 | Endophytic | Gibellulopsis nigrescens | 100 | WM-S |
| 68 | Endophytic | Penicillium sp. | 100 | WM-S |
| 78 | Endophytic | Penicillium sp. | 100 | PDM-S |
| 81 | Endophytic | Wallemia muriae | 100 | WM-S |
| 82 | Endophytic | Emericellopsis sp. | 99 | WM-S |
| 84 * | Endophytic | Hypocreales (order) | 100 | WM-S |
| 86 | Endophytic | Emericellopsis terricola | 99 | PDM-S |
| 87 * | Endophytic | Pleosporales (order) | 99 | WM-S |
| 89 | Endophytic | Trichoderma sp. | 100 | WM-S |
| 91 | Endophytic | Penicillium virgatum | 100 | GCM-S |
| 96 | Epiphytic | Penicillium brevicompactum | 100 | GYP-S |
| 104 | Epiphytic | Penicillium chrysogenum | 100 | WM-S |
| 122 | Endophytic | Penicillium brevicompactum | 100 | GCM-S |
| Strain I.D. | Taxonomic ID | Medium & Regime | IC50 Value (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HepG2 | HT29 | A375 | HCT116 | A549 | MDA-MB231 | HaCaT | |||
| 1 | Pleosporales | PDM-L | 47 | 39 | >100 | >100 | 98 | 49 | >100 |
| 35 | Cadophora malorum | PDM-L | 15 | 50 | 4 | 6 | 16 | 35 | 5 |
| 37 | Hypocreales | PDM-L | 11 | 27 | 5 | 7 | 18 | 7 | 8 |
| 50 | Penicillium sp. | PDM-L | 11 | 100 | 8 | 8 | 15 | 7 | 9 |
| PDM-S | 6 | 16 | 29 | 8 | 10 | 2 | 2 | ||
| Cza-S | >100 | >100 | 53 | 23 | 23 | 6 | 4 | ||
| 56 | Penicillium sp. | PDM-L | 43 | 36 | 66 | >100 | 42 | 35 | 69 |
| Cza-L | 100 | 100 | 63 | >100 | 40 | 32 | 32 | ||
| 58 | Fusarium graminearum | PDM-L | 74 | 69 | 72 | 74 | 85 | 78 | 78 |
| PDM-S | 69 | 63 | 59 | 69 | 71 | 62 | 44 | ||
| Cza-S | 65 | 79 | 52 | 65 | 74 | 74 | >100 | ||
| 59 | Glomerellales | PDM-S | >100 | >100 | 42 | 32 | 83 | 14 | 30 |
| 68 | Penicillium sp. | PDM-L | 10 | 32 | 8 | 7 | 10 | 5 | 8 |
| PDM-S | 12 | 18 | 28 | 6 | 9 | 3 | 3 | ||
| 78 | Penicillium sp. | PDM-L | 17 | 21 | 15 | 25 | 19 | 9 | 10 |
| 87 | Pleosporales | PDM-L | 43 | 35 | 56 | 91 | 36 | 30 | 60 |
| Stand. | 14.9 | 3 | 0.13 | 10.6 | 31.4 | 15.2 | 10 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, B.; Parrot, D.; Blümel, M.; Labes, A.; Tasdemir, D. Influence of OSMAC-Based Cultivation in Metabolome and Anticancer Activity of Fungi Associated with the Brown Alga Fucus vesiculosus. Mar. Drugs 2019, 17, 67. https://doi.org/10.3390/md17010067
Fan B, Parrot D, Blümel M, Labes A, Tasdemir D. Influence of OSMAC-Based Cultivation in Metabolome and Anticancer Activity of Fungi Associated with the Brown Alga Fucus vesiculosus. Marine Drugs. 2019; 17(1):67. https://doi.org/10.3390/md17010067
Chicago/Turabian StyleFan, Bicheng, Delphine Parrot, Martina Blümel, Antje Labes, and Deniz Tasdemir. 2019. "Influence of OSMAC-Based Cultivation in Metabolome and Anticancer Activity of Fungi Associated with the Brown Alga Fucus vesiculosus" Marine Drugs 17, no. 1: 67. https://doi.org/10.3390/md17010067
APA StyleFan, B., Parrot, D., Blümel, M., Labes, A., & Tasdemir, D. (2019). Influence of OSMAC-Based Cultivation in Metabolome and Anticancer Activity of Fungi Associated with the Brown Alga Fucus vesiculosus. Marine Drugs, 17(1), 67. https://doi.org/10.3390/md17010067