Paenidigyamycin A, Potent Antiparasitic Imidazole Alkaloid from the Ghanaian Paenibacillus sp. DE2SH
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sediment Sample Collection Sites
2.2. Taxonomy of Strain DE2SH
2.3. Structure Determination of Paenidigyamycin A (1)
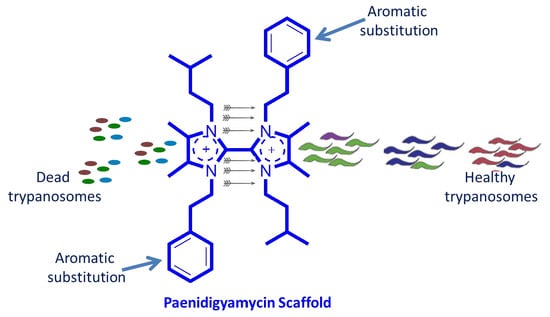
2.4. Antiparasitic and Antibacterial Activity of Paenidigyamycin A
2.5. Possible Biosynthesis of Paenidigyamycin A (1)
3. Experimental Section
3.1. General Experimental Procedures
3.2. Identification of Strain DE2SH
3.3. Fermentation
3.4. Extraction and Purification
3.5. Bioassay Reagents
3.6. Compound Preparation for Bioassay
3.7. Cell Culture
3.8. Preparation of Cercarial Suspension
3.9. Bio-Assays
3.9.1. Screening for Anti-Malaria Activity Using the SYBR Green I Assay
3.9.2. In Vitro Viability Test for Trypanosome Parasites
3.9.3. In Vitro Viability Test for Leishmania Parasites
3.9.4. In Vitro Cercariacidal Activity Test
3.9.5. In-Vitro Susceptibility Testing of Trichomonas Mobilensis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mitra, A.K.; Mawson, A.R. Neglected tropical diseases: Epidemiology and global burden. Trop. Med. Infect. Dis. 2017, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Hailu, A.; Dagne, D.A.; Boelaert, M. Leishmaniasis. In Neglected Tropical Diseases-Sub-Saharan Africa; Springer International Publishing: Cham, Switzerland, 2016; pp. 87–112. [Google Scholar]
- Flint, M.; Du Plessis, S.S. Trichomonas vaginalis in sub-Saharan Africa: Occurrence and diagnostic approaches for the male partner. Med. Technol. SA 2013, 27, 26–28. [Google Scholar]
- Cable, J.; Barber, I.; Boag, B.; Ellison, A.R.; Morgan, E.R.; Murray, K.; Emily, L.P.; Steven, M.S.; Anthony, J.W.; Booth, M. Global change, parasite transmission and disease control: Lessons from ecology. Philos. Trans. R. Soc. B 2017, 372, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Johnston, K.L.; Ford, L.; Taylor, M.J. Development and validation of a high-throughput anti-wolbachia whole-cell screen. J. Biomol. Screen 2014, 19, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Hotez, P.J.; Pecoul, B.; Rijal, S.; Boehme, C.; Aksoy, S.; Malecela, M.; Tapia-Conyer, R.; Reeder, J.C. Eliminating the neglected tropical diseases: Translational science and new technologies. PLoS Negl. Trop. Dis. 2016, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Goupil, L.S.; McKerrow, J.H. Introduction: Drug discovery and development for neglected diseases. Chem. Rev. 2014, 114, 11131–11137. [Google Scholar] [CrossRef] [PubMed]
- Ayoubi, S.; Mirtajani, S.B.; Zahiri, R.; Pourzeinolabedin, F.; Aghajani, J. A simple overview of common parasitic diseases: Which parasitic disease is more dangerous? J. Microbiol. Exp. 2017, 5, 00172. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Chai, J.Y. Praziquantel treatment in trematode and cestode infections: An update. Infect. Chemother. 2013, 45, 32–43. [Google Scholar] [CrossRef]
- Ojurongbe, O.; Sina-Agbaje, O.R.; Busari, A.; Okorie, P.N.; Ojurongbe, T.A.; Akindele, A.A. Efficacy of praziquantel in the treatment of Schistosoma haematobium infection among school-age children in rural communities of Abeokuta, Nigeria. Infect. Dis. Poverty 2014, 3, 30. [Google Scholar] [CrossRef] [Green Version]
- Wood, C.L.; Johnson, P.T. A world without parasites: Exploring the hidden ecology of infection. Front. Ecol. Environ. 2015, 13, 425–434. [Google Scholar] [CrossRef]
- Camejo, A. Parasite Killers. Trends Parasitol. 2017, 33, 151–152. [Google Scholar] [CrossRef]
- Shen, B. A New Golden Age of Natural Products Drug Discovery. Cell 2015, 163, 1297–1300. [Google Scholar] [CrossRef]
- Huang, T.; Lin, S. Microbial Natural Products: A Promising Source for Drug Discovery. J. Appl. Microbiol. Biochem. 2017, 1, 5. [Google Scholar] [CrossRef]
- Newman, D. Screening and identification of novel biologically active natural compounds. F1000Research 2017, 6. [Google Scholar] [CrossRef]
- Dalmaso, G.Z.L.; Ferreira, D.; Vermelho, A.B. Marine Extremophiles: A Source of Hydrolases for Biotechnological Applications. Marine Drugs 2015, 13, 1925–1965. [Google Scholar] [CrossRef] [Green Version]
- Coker, J.A. Extremophiles and biotechnology: Current uses and prospects. F1000Research 2016, 5. [Google Scholar] [CrossRef]
- Olalekan, E.I.; Kies, F.; Omolara, A.L.-A.; Rashidat, S.D.; Hakeem, F.B.; Latunji, A.S.; Zaid, A.A.; Emeka, N.; Charles, O.I.; Oluwaseun, F. Effect of Water Quality Characteristics on Fish Population of the Lake Volta, Ghana. J. Environ. Anal. Toxicol. 2015, 5, 1–5. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C.R. Evolution of Protein Molecules. In Mammalian, Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Parsimony in Systematics: Biological and Statistical Issues. Ann. Rev. Ecol. Syst. 1983, 14, 313–333. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An approach using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Kluge, A.G.; Farris, J.S. Quantitative phyletics and the evolution of anurans. Syst. Biol. 1969, 18, 1–32. [Google Scholar] [CrossRef]
- Keiser, J.; N’Guessan, N.A.; Adoubryn, K.D.; Silue, K.D.; Vounatsou, P.; Hatz, C.; Utzinger, J.; N’Goran, E.K. Efficacy and safety of mefloquine, artesunate, mefloquine-artesunate, and praziquantel against Schistosoma haematobium: Randomized, exploratory open-label trial. Clin. Infect Dis. 2010, 50, 1205–1213. [Google Scholar] [CrossRef]
- Kupchan, M.S.; Britton, R.W.; Zeigler, M.F.; Sigel, C.W. Bruceantin, a new potent antileukemic simaroubolide from Brucea antidysenterica. J. Org. Chem. 1973, 38, 178–179. [Google Scholar] [CrossRef]
- Yabu, Y.; Minagawab, N.; Kitac, K.; Nagaid, K.; Honmab, M.; Sakajob, S.; Koide, T.; Ohta, N.; Yoshimoto, A. Oral and intraperitoneal treatment of Trypanosoma brucei brucei with a combination of ascofuranone and glycerol in mice. Parasitol. Int. 1998, 47, 131–137. [Google Scholar] [CrossRef]
- Smilkstein, M.; Sriwilaijaroen, N.; Kelly, J.X.; Wilairat, P.; Riscoe, M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother. 2004, 48, 50–55. [Google Scholar] [CrossRef]
- Trager, W.; Jensen, J. Human malaria parasites in continuous culture. Science 1976, 193, 673–675. [Google Scholar] [CrossRef]
- Holtfreter, M.C.; Loebermann, M.; Klammt, S.; Sombetzki, M.; Bodammer, P.; Riebold, D.; Kinzelbach, R.; Reisinger, E.C. Schistosoma mansoni: Schistosomicidal effect of mefloquine and primaquine in vitro. Exp. Parasitol. 2011, 127, 270–276. [Google Scholar] [CrossRef]
- Eissa, M.M.; El Bardicy, S.; Tadros, M. Bioactivity of miltefosine against aquatic stages of Schistosoma mansoni, Schistosoma haematobium and their snail hosts, supported by scanning electron microscopy. Parasit. Vectors. 2011, 4, 73. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Peng, X.M.; Damu, G.L.; Geng, R.X.; Zhou, C.H. Comprehensive review in current developments of imidazole-based medicinal chemistry. Med. Res. Rev. 2014, 34, 340–437. [Google Scholar] [CrossRef]





| # | δC mult | δH mult (J Hz) | HMBC | # | δC mult | δH mult (J Hz) | HMBC |
|---|---|---|---|---|---|---|---|
| 1-N | 1′-N | ||||||
| 2 | 135.4, C | 8, 10 | 2′ | 135.5, C | 8′, 10′ | ||
| 3-N | 3′-N | ||||||
| 4 | 128.4, C | 10, 6 | 4′ | 128.5, C | 10′, 6′ | ||
| 5 | 128.4, C | 8, 7 | 5′ | 128.5, C | 8′, 7′ | ||
| 6 | 8.1, CH3 | 2.27, s | 6′ | 8.2, CH3 | 2.31, s | ||
| 7 | 8.1, CH3 | 2.22, s | 7′ | 8.0, CH3 | 2.06, s | ||
| 8 | 49.7, CH2 | 4.40, t (6.8) | 9 | 8′ | 49.2, CH2 | 4.33, t (6.9) | 9′ |
| 9 | 36.7, CH2 | 3.12, t (6.7) | 8, 2′, 6′ | 9′ | 37.1, CH2 | 3.04, t (6.9) | 8′ |
| 10 | 46.4, CH2 | 4.03, m | 12, 11 | 10′ | 46.6, CH2 | 4.13, m | 12′, 11′ |
| 11 | 39.5, CH2 | 1.57, m | 13, 14, 10, 12 | 11′ | 39.5, CH2 | 1.73, m | 13′, 14′, 12′, 10′ |
| 12 | 26.5, CH | 1.48, n (6.8) | 13, 14, 10 | 12′ | 26.9, CH | 1.67, n (6.8) | 13′, 14′, 11′, 10′ |
| 13 | 22.5, CH3 | 0.96, d (6.6) | 14 | 13′ | 22.5, CH3 | 1.02, d (6.5) | 14′ |
| 14 | 22.5, CH3 | 0.96, d (6.6) | 13 | 14′ | 22.5, CH3 | 1.02, d (6.5) | 13′ |
| 1′ | 137.8, C | 9, 8, 3′, 5′ | 1″ | 137.8, C | 3″, 5″, 9′, 8′ | ||
| 2′ | 130.0, CH | 7.10, m | 9, 4′, 3′, 5′ | 2″ | 130.0, CH | 7.10, m | 4″, 3″, 5″, 9′ |
| 3′ | 130.0, CH | 7.31, m | 2′, 6′ | 3″ | 130.0, CH | 7.31, m | 2″, 6″ |
| 4′ | 128.4, CH | 7.29, m | 2′, 6′ | 4″ | 128.4, CH | 7.29, m | 2″, 6″ |
| 5′ | 130.0, CH | 7.31, m | 2′, 6′ | 5″ | 130.0, CH | 7.31, m | 2″, 6″, |
| 6′ | 130.0, CH | 7.10, m | 9, 4′, 3′, 5′ | 6″ | 130.0, CH | 7.10, m | 4″, 3″, 5″, 9′ |
| Compound (1) | Microbe | IC50 (µM) | Positive Control | IC50 (µM) |
|---|---|---|---|---|
| Paenidigyamycin A | E. coli | 76.98 | Ampicillin | 10.44 |
| S. aureus | 11.87 | Ampicillin | 0.18 | |
| B. cereus | 97.23 | Ampicillin | 1.70 | |
| S. flexneri | 56.98 | Ampicillin | 1.76 | |
| S. paratyphi B | 57.98 | Ampicillin | 1.53 | |
| L. monocytogenes | 18.98 | Ampicillin | 2.71 |
| Compound (1) | Parasite | IC50 (µM) | Positive Control | IC50 |
|---|---|---|---|---|
| Paenidigyamycin A | P. falciparum 3d7 | 9.08 | Artesunate | 36 nm |
| T. brucei brucei | 0.78 | Coptis japonica | 8.20 µM | |
| L. donovani | 7.02 | Amphotericin B | 0.32 µM | |
| L. major | 0.75 | Amphotericin B | 0.31 µM |
| Compound (1) | Parasite | IC50 (µM) | Positive Control | IC50 (µM) |
|---|---|---|---|---|
| Paenidigyamycin A | T. mobilensis | > 100 | Metronidazole | 5.2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osei, E.; Kwain, S.; Mawuli, G.T.; Anang, A.K.; Owusu, K.B.-A.; Camas, M.; Camas, A.S.; Ohashi, M.; Alexandru-Crivac, C.-N.; Deng, H.; et al. Paenidigyamycin A, Potent Antiparasitic Imidazole Alkaloid from the Ghanaian Paenibacillus sp. DE2SH. Mar. Drugs 2019, 17, 9. https://doi.org/10.3390/md17010009
Osei E, Kwain S, Mawuli GT, Anang AK, Owusu KB-A, Camas M, Camas AS, Ohashi M, Alexandru-Crivac C-N, Deng H, et al. Paenidigyamycin A, Potent Antiparasitic Imidazole Alkaloid from the Ghanaian Paenibacillus sp. DE2SH. Marine Drugs. 2019; 17(1):9. https://doi.org/10.3390/md17010009
Chicago/Turabian StyleOsei, Enoch, Samuel Kwain, Gilbert Tetevi Mawuli, Abraham Kwabena Anang, Kofi Baffour-Awuah Owusu, Mustafa Camas, Anil Sazak Camas, Mitsuko Ohashi, Cristina-Nicoleta Alexandru-Crivac, Hai Deng, and et al. 2019. "Paenidigyamycin A, Potent Antiparasitic Imidazole Alkaloid from the Ghanaian Paenibacillus sp. DE2SH" Marine Drugs 17, no. 1: 9. https://doi.org/10.3390/md17010009
APA StyleOsei, E., Kwain, S., Mawuli, G. T., Anang, A. K., Owusu, K. B. -A., Camas, M., Camas, A. S., Ohashi, M., Alexandru-Crivac, C. -N., Deng, H., Jaspars, M., & Kyeremeh, K. (2019). Paenidigyamycin A, Potent Antiparasitic Imidazole Alkaloid from the Ghanaian Paenibacillus sp. DE2SH. Marine Drugs, 17(1), 9. https://doi.org/10.3390/md17010009