Hybrid Polyketides from a Hydractinia-Associated Cladosporium sphaerospermum SW67 and Their Putative Biosynthetic Origin
Abstract
:1. Introduction
2. Results and Discussions
2.1. Isolation and Structural Characterization
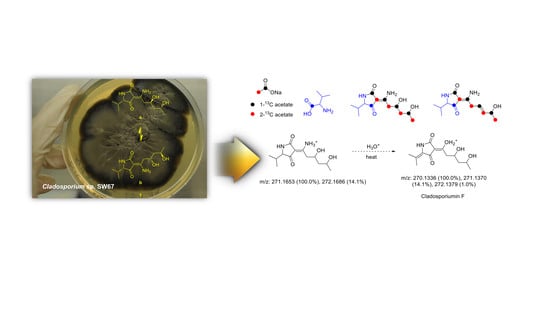
2.2. Proposed Biogenesis of 1–4
2.3. Biological Evaluation of 1–4
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Biological Material
3.3. Extraction and Isolation
Cladosin L (1)
3.4. Preparation of Mosher Ester Derivatives from Cladosin L (1)
3.5. Computational Analysis
3.6. Isotope Labeling
3.7. Renoprotective Effects against Cisplatin-Induced Kidney Cell Damage
3.8. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shnit-Orland, M.; Kushmaro, A. Coral mucus-associated bacteria: A possible first line of defense. FEMS Microbiol. Ecol. 2009, 67, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.B.; Kellogg, C.A. Microbial ecology of corals, sponges, and algae in mesophotic coral environments. FEMS Microbiol. Ecol. 2010, 73, 17–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piel, J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2004, 21, 519–538. [Google Scholar] [CrossRef] [PubMed]
- Rohwer, F.; Seguritan, V.; Azam, F.; Knowlton, N. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 2002, 243, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Lan, W.J.; Fu, S.J.; Xu, M.Y.; Liang, W.L.; Lam, C.K.; Zhong, G.H.; Xu, J.; Yang, D.P.; Li, H.J. Five new cytotoxic metabolites from the marine fungus Neosartorya Pseudofischeri. Mar. Drugs 2016, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Guo, L.; Hao, J.; Wang, L.; Zhu, W. α-glucosidase inhibitors from the marine-derived fungus Aspergillus flavipes HN4-13. J. Nat. Prod. 2016, 79, 2977–2981. [Google Scholar] [CrossRef] [PubMed]
- Julianti, E.; Oh, H.; Jang, K.H.; Lee, J.K.; Lee, S.K.; Oh, D.C. Acremostrictin, a highly oxygenated metabolite from the marine fungus Acremonium strictum. J. Nat. Prod. 2011, 74, 2592–2594. [Google Scholar] [CrossRef]
- Abdel-Lateff, A.; König, G.M.; Fisch, K.M.; Höller, U.; Jones, P.G.; Wright, A.D. New antioxidant hydroquinone derivatives from the algicolous marine fungus Acremonium sp. J. Nat. Prod. 2002, 65, 1605–1611. [Google Scholar] [CrossRef]
- Kim, K.S.; Cui, X.; Lee, D.S.; Sohn, J.H.; Yim, J.H.; Kim, Y.C.; Oh, H. Anti-inflammatory effect of neoechinulin A from the marine fungus Eurotium sp. SF-5989 through the suppression of NF-κB and p38 MAPK pathways in lipopolysaccharide-stimulated RAW264.7 macrophages. Molecules 2013, 18, 13245–13259. [Google Scholar] [CrossRef]
- Ohkawa, Y.; Miki, K.; Suzuki, T.; Nishio, K.; Sugita, T.; Kinoshita, K.; Takahashi, K.; Koyama, K. Antiangiogenic metabolites from a marine-derived fungus, Hypocrea vinosa. J. Nat. Prod. 2010, 73, 579–582. [Google Scholar] [CrossRef]
- Lee, D.S.; Ko, W.; Quang, T.H.; Kim, K.S.; Sohn, J.H.; Jang, J.H.; Ahn, J.S.; Kim, Y.C.; Oh, H. Penicillinolide A: A new anti-inflammatory metabolites from the marine fungus Penicillium sp. SF-5292. Mar. Drugs 2013, 11, 4510–4526. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.C.; Gunasekera, S.P.; Lopez, J.V.; McCarthy, P.J.; Wright, A.E. Metabolites from the marine-derived fungus Chromocleista sp. Isolated from a deep-water sediment sample collected in the gulf of Mexico. J. Nat. Prod. 2006, 69, 580–584. [Google Scholar] [CrossRef] [PubMed]
- So, H.M.; Eom, H.J.; Lee, D.; Kim, S.; Kang, K.S.; Lee, I.K.; Baek, K.H.; Park, J.Y.; Kim, K.H. Bioactivity evaluations of betulin identified from the bark of Betula platyphylla var. japonica for cancer therapy. Arch. Pharm. Res. 2018, 41, 815–822. [Google Scholar] [PubMed]
- Yu, J.S.; Roh, H.S.; Baek, K.H.; Lee, S.; Kim, S.; So, H.M.; Moon, E.; Pang, C.; Jang, T.S.; Kim, K.H. Bioactivity-guided isolation of ginsenosides from Korean Red Ginseng with cytotoxic activity against human lung adenocarcinoma cells. J. Ginseng Res. 2018, 42, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.C.; Choi, E.; Eom, H.J.; Jo, M.S.; Kim, S.; So, H.M.; Kim, S.H.; Kang, K.S.; Kim, K.H. LC/MS-based analysis of bioactive compounds from the bark of Betula platyphylla var. japonica and Their Effects on Regulation of Adipocyte and Osteoblast Differentiation. Nat. Prod. Sci. 2018, 24, 235–240. [Google Scholar]
- Lee, S.R.; Park, Y.J.; Han, Y.B.; Lee, J.C.; Lee, S.; Park, H.J.; Lee, H.J.; Kim, K.H. Isoamericanoic acid B from Acer tegmentosum as a potential phytoestrogen. Nutrients 2018, 10, 1915. [Google Scholar] [CrossRef]
- Kang, H.R.; Yun, H.S.; Lee, T.K.; Lee, S.; Kim, S.H.; Moon, E.; Park, K.M.; Kim, K.H. Chemical characterization of novel natural products from the roots of Asian rice (Oryza sativa) that control adipocyte and osteoblast differentiation. J. Agric. Food Chem. 2018, 66, 2677–2684. [Google Scholar] [CrossRef]
- Rischer, M.; Lee, S.R.; Eom, H.J.; Park, H.B.; Vollmers, J.; Kaster, A.K.; Shin, Y.H.; Oh, D.C.; Kim, K.H.; Beemelmanns, C. Spirocyclic cladosporicin A and cladosporiumins I and J from a Hydractinia-associated Cladosporium sphaerospermum SW67. Org. Chem. Front. 2019, 6, 1084–1093. [Google Scholar] [CrossRef]
- Wu, G.; Sun, X.; Yu, G.; Wang, W.; Zhu, T.; Gu, Q.; Li, D. Cladosins A−E, Hybrid polyketides from a deep-sea-derived fungus, Cladosporium sphaerospermum. J. Nat. Prod. 2014, 77, 270–275. [Google Scholar] [CrossRef]
- Zhang, Z.; He, X.; Wu, G.; Liu, C.; Lu, C.; Gu, Q.; Che, Q.; Zhu, T.; Zhang, G.; Li, D. Aniline-tetramic acids from the deep-sea-derived fungus Cladosporium sphaerospermum L3P3 cultured with the HDAC Inhibitor SAHA. J. Nat. Prod. 2018, 81, 1651–1657. [Google Scholar] [CrossRef]
- Jeong, Y.C.; Moloney, M.G. Synthesis of and tautomerism in 3-acyltetramic acids. J. Org. Chem. 2011, 76, 1342–1354. [Google Scholar] [CrossRef] [PubMed]
- Aoki, S.; Higuchi, K.; Ye, Y.; Satari, R.; Kobayashi, M. Melophlins A and B, novel tetramic acids reversing the phenotype of ras-transformed cells, from the marine sponge Melophlus sarassinorum. Tetrahedron 2000, 56, 1833–1836. [Google Scholar] [CrossRef]
- Liang, X.; Huang, Z.H.; Ma, X.; Qi, S.H. Unstable tetramic acid derivatives from deep-sea-derived fungus Cladosporium sphaerospermum EIODSF 008. Mar. Drugs 2018, 16, 448. [Google Scholar] [CrossRef] [PubMed]
- Freire, F.; Seco, J.M.; Quinoa, E.; Riguera, R. Determining the absolute stereochemistry of secondary/secondary diols by 1H NMR: Basis and applications. J. Org. Chem. 2005, 70, 3778–3790. [Google Scholar] [CrossRef]
- Yu, G.H.; Wu, G.W.; Zhu, T.J.; Gu, Q.Q.; Li, D.H. Cladosins F and G, two new hybrid polyketides from the deep-sea-derived Cladosporium sphaerospermum 2005-01-E3. J. Asian Nat. Prod. Res. 2015, 17, 120–124. [Google Scholar] [CrossRef]
- Mo, X.; Li, Q.; Ju, J. Naturally occurring tetramic acid products: Isolation, structure elucidation and biological activity. RSC Adv. 2014, 4, 50566–50593. [Google Scholar] [CrossRef]
- Fisch, K.M. Biosynthesis of natural products by microbial iterative hybrid PKS–NRPS. RSC Adv. 2013, 3, 18228–18247. [Google Scholar] [CrossRef]
- Boettger, D.; Hertweck, C. Molecular Diversity Sculpted by Fungal PKS–NRPS Hybrids. ChemBioChem 2013, 14, 28–42. [Google Scholar] [CrossRef]
- Miyanaga, A.; Kudo, F.; Eguchi, T. Protein–protein interactions in polyketide synthase–nonribosomal peptide synthetase hybrid assembly lines. Nat. Prod. Rep. 2018, 35, 1185–1209. [Google Scholar] [CrossRef]
- Du, L.; Lou, L. PKS and NRPS release mechanisms. Nat. Prod. Rev. 2010, 27, 255–278. [Google Scholar] [CrossRef]
- Bachmann, B.O.; Ravel, J. Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009, 458, 181–217. [Google Scholar] [PubMed]
- BLAST: Basic Local Alignment Search Tool—NCBI-NIH. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 7 October 2019).
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updatest o the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- MIBiG: Minimum Information about a Biosynthetic Gene Cluster. Available online: https://mibig.secondarymetabolites.org/ (accessed on 7 October 2019).
- Online Resource of Protein Sequence and Functional Information. Available online: https://www.uniprot.org/ (accessed on 7 October 2019).
- Hall, T.A. Bioedit: A user-friendly biological sequences alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Li, Y.; Dodge, G.J.; Fiers, W.D.; Fecik, R.A.; Smith, J.L.; Aldrich, C.C. Functional characterization of a dehydratase domain from the pikromycin polyketide synthase. J. Am. Chem. Soc. 2015, 137, 7003–7006. [Google Scholar] [CrossRef]
- Kagan, R.M.; Clarke, S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 1994, 310, 417–427. [Google Scholar] [CrossRef]
- Ansari, M.Z.; Sharma, J.; Gokhale, R.S.; Mohanty, D. In silico analysis of methyltransferase domains involved in biosynthesis of secondary metabolites. BMC Bioinform. 2008, 9, 454. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, Z.; Shao, C.-L.; Wang, C.-Y. Analysis of the Sequences, Structures, and Functions of Product-Releasing Enzyme Domains in Fungal Polyketide Synthases. Front. Microbiol. 2017, 8, 1685. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Eley, K.L.; Halo, L.M.; Song, Z.; Powles, H.; Cox, R.J.; Bailey, A.M.; Lazarus, C.M.; Simpson, T.J. Biosynthesis of the 2-pyridone tenellin in the insect pathogenic fungus Beauveria bassiana. ChemBioChem 2007, 8, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Halo, L.M.; Marshall, J.W.; Yakasai, A.A.; Song, Z.; Butts, C.P.; Crump, M.P.; Heneghan, M.; Bailey, A.M.; Simpson, T.J.; Lazarus, C.M.; et al. Authentic heterologous expression of the tenellin iterative polyketide synthase nonribosomal peptide synthetase requires coexpression with an enoyl reductase. ChemBioChem 2008, 9, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, S.; Schumann, J.; Scherlach, K.; Lange, C.; Brakhage, A.A.; Hertweck, C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 2007, 3, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Gui, C.; Li, Q.; Mo, X.; Qin, X.; Ma, J.; Ju, J. Discovery of a New Family of Dieckmann Cyclases Essential to Tetramic Acid and Pyridone-Based Natural Products Biosynthesis. Org. Lett. 2015, 17, 628–631. [Google Scholar] [CrossRef] [PubMed]
- Greule, A.; Stok, J.E.; De Voss, J.J.; Cryle, M.J. Unrivalled diversity: The many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat. Prod. Rep. 2018, 35, 757–791. [Google Scholar] [CrossRef]
- Lee, D.; Kang, K.S.; Lee, H.J.; Kim, K.H. Chemical characterization of a renoprotective metabolite from termite-associated Streptomyces sp. RB1 against cisplatin-induced cytotoxicity. Int. J. Mol. Sci. 2018, 19, 174. [Google Scholar] [CrossRef]
- Lee, D.; Lee, S.; Shim, S.H.; Lee, H.J.; Choi, Y.; Jang, T.S.; Kim, K.H.; Kang, K.S. Protective effect of lanostane triterpenoids from the sclerotia of Poria cocos Wolf against cisplatin-induced apoptosis in LLC-PK1 cells. Bioorg. Med. Chem. Lett. 2017, 27, 2881–2885. [Google Scholar] [CrossRef]
- Lee, S.R.; Lee, D.; Lee, H.J.; Noh, H.J.; Jung, K.; Kang, K.S.; Kim, K.H. Renoprotective chemical constituents from an edible mushroom, Pleurotus cornucopiae in cisplatin-induced nephrotoxicity. Bioorg. Chem. 2017, 71, 67–73. [Google Scholar] [CrossRef]
- Volarevic, V.; Djokovic, B.; Jankovic, M.G.; Harrell, C.R.; Fellabaum, C.; Djonov, V.; Arsenijevic, N. Molecular mechanisms of cisplatin-induced nephrotoxicity: A balance on the knife edge between renoprotection and tumor toxicity. J. Biomed. Sci. 2019, 26, 25. [Google Scholar] [CrossRef]
- Oh, G.S.; Kim, H.J.; Shen, A.; Lee, S.B.; Khadka, D.; Pandit, A.; So, H.S. Cisplatin-induced kidney dysfunction and perspectives on improving treatment strategies. Electrolyte Blood Press 2014, 12, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Panichpisal, K.; Kurtzman, N.; Nugent, K. Cisplatin nephrotoxicity: A review. Am. J. Med. Sci. 2007, 334, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Ojha, S.; Venkataraman, B.; Kurdi, A.; Mahgoub, E.; Sadek, B.; Rajesh, M. Plant-derived agents for counteracting cisplatin-induced nephrotoxicity. Oxid. Med. Cell Longev. 2016, 2016, 4320374. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tao, X.; Yin, L.; Xu, L.; Xu, Y.; Qi, Y.; Han, X.; Song, S.; Zhao, Y.; Lin, Y.; et al. Protective effects of dioscin against cisplatin-induced nephrotoxicity via the microRNA-34a/sirtuin 1 signalling pathway. Br. J. Pharmacol. 2017, 174, 2512–2527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruhn, T.; Schaumlöffel, A.; Hemberger, Y.; Pescitelli, G. SpecDis, Version 1.70.1, Berlin, Germany. 2017. Available online: https://specdis-software.jimdo.com (accessed on 7 October 2019).

 ) and HMBC (→) correlations for 1.
) and HMBC (→) correlations for 1.






| Position | 1 | |||
|---|---|---|---|---|
| Exo-Form A (a) | Exo-Form B (b) | |||
| δH (J in Hz) | δC, typ | δH (J in Hz) | δC, typ | |
| 2 | 178.3, CO | 176.5, CO | ||
| 3 | 99.1, C | 97.7, C | ||
| 4 | 199.8, CO | 202.0, CO | ||
| 5 | 3.59, d (3.0) | 68.0, CH | 3.66 d (3.0) | 67.0, CH |
| 6 | 172.3, C | 172.7, C | ||
| 7 | 2.85, m; 3.17, dd (13.0, 4.0) | 41.1, CH2 | 2.82 m; 3.13 dd (13.5, 4.5) | 42.1, CH2 |
| 8 | 4.14, m | 69.6, CH | 4.13 m | 69.2, CH |
| 9 | 1.59, m; 1.60, m | 48.0, CH2 | 1.59 m; 1.60 m | 47.8, CH2 |
| 10 | 3.99, m | 66.1, CH | 3.99 m | 66.1, CH |
| 11 | 1.18, d (6.0) | 25.0, CH3 | 1.18 d (6.0) | 25.1, CH3 |
| 12 | 2.13, m | 32.1, CH | 2.12 m | 32.1, CH |
| 13 | 0.77, d (7.0) | 16.4, CH3 | 0.78 d (7.0) | 16.7, CH3 |
| 14 | 1.01, d (7.0) | 20.7, CH3 | 1.01 d (7.0) | 20.6, CH3 |
| Amino Acid Sequence | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SW67_clsI_unknown_1 | R | E | W | E | T | Q | F | Q | L | T | P | M | A | A | D | S | R | Y | N | F | R | L | M | I | C | G | P | S | E | S |
| UM843_cluI_unknown_1 | R | E | W | E | T | Q | F | Q | L | T | P | M | A | A | D | S | R | Y | N | F | R | L | M | I | C | G | P | S | E | S |
| ACLA_078660_DH | F | N | H | S | Q | P | L | L | I | H | P | A | T | L | D | A | A | I | Q | S | I | M | L | A | Y | C | Y | P | G | D |
| A0A0C6E017_DH | D | M | Q | I | D | N | Y | V | V | N | P | G | F | L | D | V | A | F | Q | S | V | Y | T | A | F | S | S | P | A | S |
| ANIA_08412_DH | V | P | D | A | D | E | L | L | V | H | P | I | D | L | D | A | A | F | Q | S | V | M | L | A | Y | S | Y | P | G | D |
| A0A0C6E0I7_DH | P | V | S | W | T | H | T | L | T | H | P | A | P | I | D | T | A | V | Q | G | L | L | T | A | F | S | F | P | G | D |
| FFUJ_12239_DH | F | N | H | S | Q | P | L | L | I | H | P | A | T | L | D | A | A | I | Q | S | I | M | L | A | Y | C | Y | P | G | D |
| ACLA_078660_DH | C | L | S | D | T | G | L | L | V | H | P | A | F | L | D | M | T | L | H | A | T | L | A | A | F | A | S | P | G | D |
| B1GVX7_DH | P | V | S | W | T | H | T | L | T | H | P | A | P | I | D | T | A | V | Q | G | L | L | T | A | F | S | F | P | G | D |
| A0JJU1_DH | A | D | L | N | D | C | Y | L | V | H | P | A | I | L | D | V | A | F | Q | T | I | F | V | A | R | A | H | P | D | S |
| S0EET5_DH | V | V | P | D | F | P | A | M | I | H | P | A | L | I | D | G | A | F | Q | S | I | F | A | A | Y | C | Q | P | D | D |
| Amino Acid Sequence | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SW67_clsI_unknown_2 | - | - | - | - | - | - | - | F | D | Q | E | N | H | R | V | S | P | G | G | C | I | C | V | L | H | S | R | T | - | - |
| UM843_cluI_unknown_2 | - | - | - | - | - | - | - | F | D | Q | E | N | H | R | V | S | P | G | G | C | I | C | V | L | H | S | R | T | - | - |
| ACLA_078660_C-MT | T | R | D | L | A | Q | T | V | R | N | V | R | R | L | L | K | P | G | G | Y | L | L | L | L | E | I | T | E | N | - |
| A0A0C6E017_C-MT | C | A | R | L | D | E | A | V | A | N | L | R | K | L | L | K | P | G | G | L | L | V | L | G | E | G | A | S | D | G |
| ANIA_08412_C-MT | T | H | S | L | E | N | T | L | R | Q | C | R | K | L | L | R | P | G | G | R | L | V | L | L | E | I | T | R | - | - |
| A0A0C6E0I7_C-MT | T | R | D | L | A | Q | T | V | R | N | V | R | R | L | L | K | P | G | G | Y | L | L | L | L | E | I | T | E | N | - |
| FFUJ_12239_C-MT | T | E | F | L | E | K | T | M | R | N | V | R | T | L | L | K | P | G | G | Y | L | C | L | L | E | C | T | G | - | - |
| ACLA_078660_C-MT | T | H | S | L | E | N | T | L | R | Q | C | R | K | L | L | R | P | G | G | R | L | V | L | L | E | I | T | R | - | - |
| B1GVX7_C-MT | T | R | N | L | G | V | T | L | G | N | V | R | S | L | L | K | P | G | G | Y | L | L | L | N | E | K | T | G | P | - |
| A0JJU1_C-MT | T | K | S | L | T | V | T | M | R | N | T | R | K | L | L | K | P | G | G | Q | L | L | L | L | E | V | T | S | - | - |
| S0EET5_C-MT | T | P | D | L | E | K | T | M | A | H | A | R | S | L | L | K | P | G | G | Q | M | V | I | L | E | I | T | H | K | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.R.; Lee, D.; Eom, H.J.; Rischer, M.; Ko, Y.-J.; Kang, K.S.; Kim, C.S.; Beemelmanns, C.; Kim, K.H. Hybrid Polyketides from a Hydractinia-Associated Cladosporium sphaerospermum SW67 and Their Putative Biosynthetic Origin. Mar. Drugs 2019, 17, 606. https://doi.org/10.3390/md17110606
Lee SR, Lee D, Eom HJ, Rischer M, Ko Y-J, Kang KS, Kim CS, Beemelmanns C, Kim KH. Hybrid Polyketides from a Hydractinia-Associated Cladosporium sphaerospermum SW67 and Their Putative Biosynthetic Origin. Marine Drugs. 2019; 17(11):606. https://doi.org/10.3390/md17110606
Chicago/Turabian StyleLee, Seoung Rak, Dahae Lee, Hee Jeong Eom, Maja Rischer, Yoon-Joo Ko, Ki Sung Kang, Chung Sub Kim, Christine Beemelmanns, and Ki Hyun Kim. 2019. "Hybrid Polyketides from a Hydractinia-Associated Cladosporium sphaerospermum SW67 and Their Putative Biosynthetic Origin" Marine Drugs 17, no. 11: 606. https://doi.org/10.3390/md17110606
APA StyleLee, S. R., Lee, D., Eom, H. J., Rischer, M., Ko, Y. -J., Kang, K. S., Kim, C. S., Beemelmanns, C., & Kim, K. H. (2019). Hybrid Polyketides from a Hydractinia-Associated Cladosporium sphaerospermum SW67 and Their Putative Biosynthetic Origin. Marine Drugs, 17(11), 606. https://doi.org/10.3390/md17110606