Virescenosides from the Holothurian-Associated Fungus Acremonium striatisporum Kmm 4401
Abstract
:1. Introduction
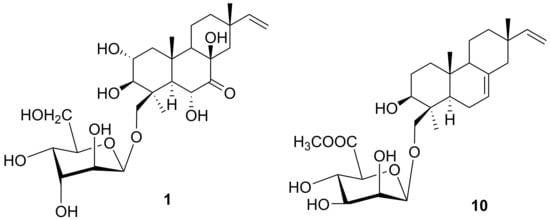
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Cultivation of Fungus
3.3. Extraction and Isolation
3.4. Spectral Data
3.5. Urease Inhibition Assay
3.6. Reactive Oxygen Species (ROS) Level Analysis in LPS-Treated Cells
3.7. Reactive Nitrogen Species (RNS) Level Analysis in LPS-Treated Cells
3.8. Peritoneal Macrophage Isolation
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Du, F.Y.; Li, X.M.; Song, J.Y.; Li, C.S.; Wang, B.G. Anthraquinone derivatives and an orsellinic acid ester from the marine alga-derived endophytic fungus Eurotium cristatum EN-220. Helv. Chim. Acta 2014, 97, 973–978. [Google Scholar] [CrossRef]
- Li, D.L.; Li, X.M.; Wang, B.G. Natural anthraquinone derivatives from a marine mangrove plant-derived endophytic fungus Eurotium rubrum: Structural elucidation and DPPH radical scavenging activity. J. Microbiol. Biotechnol. 2009, 19, 675–678. [Google Scholar]
- Li, Y.; Li, X.; Lee, U.; Kang, J.S.; Choi, H.D.; Son, B.W. A new radical scavenging anthracene glycoside, asperflavin ribofuranoside, and polyketides from a marine isolate of the fungus Microsporum. Chem. Pharm. Bull. 2006, 54, 882–883. [Google Scholar] [CrossRef]
- Li, X.F.; Xia, Z.Y.; Tang, J.Q.; Wu, J.H.; Tong, J.; Li, M.J.; Ju, J.H.; Chen, H.R.; Wang, L.Y. Identification and biological evaluation of secondary metabolites from marine derived fungi—Aspergillus sp. SCSIOW3, cultivated in the presence of epigenetic modifying agents. Molecules 2017, 22, 1302. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Mou, Y.H.; Dong, Y.; Wu, Y.; Liu, B.Y.; Bai, J.; Yan, D.J.; Zhang, L.; Feng, D.Q.; Pei, Y.H.; et al. Diphenyl ethers from a marine-derived Aspergillus sydowii. Mar. Drugs 2018, 16, 451. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.X.; Xue, Y.B.; Bi, X.B.; Zhang, J.W.; Luo, Z.W.; Li, X.N.; Yao, G.M.; Wang, J.P.; Zhang, Y.H. Five new secondary metabolites produced by a marine-associated fungus, Daldinia eschscholzii. Mar. Drugs 2014, 12, 5563–5575. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhu, T.J.; Liu, H.B.; Fang, Y.C.; Zhu, W.M.; Gu, Q.Q. Cytotoxic polyketides from a marine-derived fungus Aspergillus glaucus. J. Nat. Prod. 2008, 71, 1837–1842. [Google Scholar] [CrossRef]
- Harms, H.; Kehraus, S.; Nesaei-Mosaferan, D.; Hufendieck, P.; Meijer, L.; Konig, G.M. A beta-42 lowering agents from the marine-derived fungus Dichotomomyces cejpii. Steroids 2015, 104, 182–188. [Google Scholar] [CrossRef]
- Nazir, M.; Harms, H.; Loef, I.; Kehraus, S.; El Maddah, F.; Arslan, I.; Rempel, V.; Muller, C.E.; Konig, G.M. GPR18 inhibiting amauromine and the novel triterpene glycoside auxarthonoside from the sponge-derived fungus Auxarthron reticulatum. Planta Med. 2015, 81, 1141–1145. [Google Scholar] [CrossRef]
- Afiyatullov, S.S.; Pivkin, M.V.; Kalinovsky, A.I.; Kuznetsova, T.A. New cytotoxic glycosides of the fungus Acremonium striatisporum isolated from a sea cucumber. Nat. Prod. I 2007, 15, 85–114. [Google Scholar]
- Afiyatullov, S.S.; Kalinovsky, A.I.; Antonov, A.S.; Zhuravleva, O.I.; Khudyakova, Y.V.; Aminin, D.L.; Yurchenko, A.N.; Pivkin, M.V. Isolation and structures of virescenosides from the marine-derived fungus Acremonium striatisporum. Phytochem. Lett. 2016, 15, 66–71. [Google Scholar] [CrossRef]
- Afiyatullov, S.S.; Kalinovsky, A.I.; Antonov, A.S. New virescenosides from the marine-derived fungus Acremonium striatisporum. Nat. Prod. Commun. 2011, 6, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Polonsky, J.; Baskevitch, Z.; Cagnoli-Bellavita, N.; Ceccherelli, P. Structures des virescenols A et B, metabolites de Oospora virescens (link) Wallr. Tetrahedron 1970, 29, 449–454. [Google Scholar]
- Polonsky, J.; Baskevitch, Z.; Cagnoli-Bellavita, N.; Ceccherelli, P.; Buckwalter, B.L.; Wenkert, E. Carbon-13 nuclear magnetic resonance spectroscopy of naturally occurring substances. XI. Biosynthesis of virescenosides. J. Am. Chem. Soc. 1972, 94, 4369–4370. [Google Scholar] [CrossRef]
- Wenkert, E.; Beak, P. The stereochemistry of rumiene. J. Am. Chem. Soc. 1961, 83, 998–1000. [Google Scholar] [CrossRef]
- De Pascual, J.T.; Barrero, A.F.; Muriel, L.; San Feliciano, A.; Grande, M. New natural diterpene acid from Juniperus communis. Phytochemistry 1980, 19, 1153–1156. [Google Scholar] [CrossRef]
- Afiyatullov, S.S.; Kalinovsky, A.I.; Kuznetsova, T.A.; Isakov, V.V.; Pivkin, M.V.; Dmitrenok, P.S.; Elyakov, G.B. New diterpene glycosides of the fungus Acremonium striatisporum isolated from a sea cucumber. J. Nat. Prod. 2002, 65, 641–644. [Google Scholar] [CrossRef]
- Afiyatullov, S.S.; Kuznetsova, T.A.; Isakov, V.V.; Pivkin, M.V.; Prokof’eva, N.G.; Elyakov, G.B. New diterpenic altrosides of the fungus Acremonium striatisporum isolated from a sea cucumber. J. Nat. Prod. 2000, 63, 848–850. [Google Scholar] [CrossRef]
- Ceccherelli, P.; Cagnoli-Bellavita, N.; Polonsky, J.; Baskevitch, Z. Structures des virescenosides F et G, nouveaux metabolites de Oospora virescens (link) Wallr. Tetrahedron 1973, 29, 449–454. [Google Scholar] [CrossRef]
- Bellavita, N.; Bernassau, J.-M.; Ceccherelli, P.; Raju, M.S.; Wenkert, E. An unusual solvent dependence of the carbone-13 nuclear magnetic resonance spectral features of some glycosides as studied by relaxation-time measurements. J. Am. Chem. Soc. 1980, 2, 17–20. [Google Scholar] [CrossRef]
- Afiyatullov, S.S.; Kalinovsky, A.I.; Pivkin, M.V.; Dmitrenok, P.S.; Kuznetsova, T.A. New diterpene glycosides of the fungus Acremonium striatisporum isolated from a sea cucumber. Nat. Prod. Res. 2006, 20, 902–908. [Google Scholar] [CrossRef] [PubMed]
- King-Morris, M.J.; Serianni, A.S. 13C NMR studies of [1-13C] aldoses: Empirical rules correlating pyranose ring configuration and conformation with 13C chemical shifts and 13C-13C spin couplings. J. Am. Chem. Soc. 1987, 109, 3501–3508. [Google Scholar] [CrossRef]
- Podlasek, C.A.; Wu, J.; Stripe, W.A.; Bondo, P.B.; Serrianni, A.S. [13C]-Enriched methyl aldopyranosides: Structural interpretations of 13C‒1H spin-coupling constants and 1H chemical shifts. J. Am. Chem. Soc. 1995, 117, 8635–8644. [Google Scholar] [CrossRef]
- Burne, R.A.; Chen, Y.Y. Bacterial ureases in infectious diseases. Microbes Infect. 2000, 2, 533–542. [Google Scholar] [CrossRef]
- Mobley, H.L.; Hausinger, R.P. Microbial ureases: Significance, regulation, and molecular characterization. Microbiol. Rev. 1989, 53, 85–108. [Google Scholar]
- Weatherburn, M.W. Phenol hypochlorite reaction for determination of ammonia. Anal. Chem. 1967, 39, 971–974. [Google Scholar] [CrossRef]
- Ivanchina, N.V.; Kicha, A.A.; Malyarenko, T.V.; Kalinovsky, A.I.; Menchinskaya, E.S.; Pislyagin, E.A.; Dmitrenok, P.S. The influence on LPS-induced ROS formation in macrophages of capelloside A, a new steroid glycoside from the starfish Ogmaster capella. Nat. Prod. Commun. 2015, 10, 1937–1940. [Google Scholar] [CrossRef]




| Position | 1a | 2b | 3b | 4c | 5b | 6c | 7b | 8c | 9b | 10d |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 46.9, CH2 | 35.9, CH2 | 44.1, CH2 | 40.0, CH2 | 48.0, CH2 | 46.6, CH2 | 44.1, CH2 | 40.3, CH2 | 48.1, CH2 | 38.5, CH2 |
| 2 | 69.1, CH | 29.0, CH2 | 69.6, CH | 29.4, CH2 | 69.5, CH | 69.3, CH | 69.4, CH | 37.3, CH2 | 69.5, CH | 28.6, CH2 |
| 3 | 84.7, CH | 80.0, CH | 84.6, CH | 81.5, CH | 85.6, CH | 84.7, CH | 84.4, CH | 218.4, C | 85.6, CH | 79.0, CH |
| 4 | 43.8, C | 41.3, C | 44.7, C | 43.8, C | 44.6, C | 44.9, C | 44.7, C | 54.1, C | 44.6, C | 43.0, C |
| 5 | 55.9, CH | 51.8, CH | 51.7, CH | 53.5, CH | 53.2, CH | 51.9 CH | 51.5, CH | 55.5, CH | 53.2, CH | 51.5 CH |
| 6 | 57.3, CH | 37.3, CH2 | 37.6, CH2 | 25.2, CH2 | 25.3, CH2 | 39.3, CH2 | 37.8, CH2 | 25.8, CH2 | 25.3, CH2 | 24.6, CH2 |
| 7 | 178.0, C | 202.9, C | 202.7, C | 123.1, CH | 123.2, CH | 203.1, C | 202.8, C | 123.1, CH | 123.2, CH | 122.8, CH |
| 8 | 80.2, C | 130.3, C | 130.3, C | 137.2, C | 137.1, C | 136.8, C | 130.2, C | 137.7, C | 136.9, C | 134.9, C |
| 9 | 60.5, CH | 168.7, C | 167.8, C | 54.1, CH | 54.1, CH | 52.6, CH | 167.9, C | 53.1, CH | 54.1, CH | 52.3, CH |
| 10 | 43.8, C | 43.9, C | 42.2, C | 36.8, C | 37.9, C | 38.4, C | 42.2, C | 37.0, C | 37.9, C | 35.5, C |
| 11 | 18.4, CH2 | 24.9, CH2 | 25.0, CH2 | 22.1, CH2 | 22.2, CH2 | 21.0, CH2 | 25.0, CH2 | 22.1, CH2 | 22.2, CH2 | 20.5, CH2 |
| 12 | 34.3, CH2 | 35.3, CH2 | 35.2, CH2 | 37.9, CH2 | 37.8, CH2 | 35.6, CH2 | 35.2, CH2 | 37.8, CH2 | 37.8, CH2 | 36.4, CH2 |
| 13 | 35.8, C | 36.0, C | 36.1, C | 38.4, C | 38.4, C | 40.4, C | 36.1, C | 38.4, C | 38.4, C | 37.1, C |
| 14 | 49.8, CH2 | 34.9, CH2 | 34.9, CH2 | 47.7, CH2 | 47.6, CH2 | 146.5, CH | 34.9, CH2 | 47.6, CH2 | 47.6, CH2 | 46.3, CH2 |
| 15 | 151.0, CH | 147.0, CH | 147.2, CH | 152.0, CH | 151.9, CH | 148.6, CH | 146.9, CH | 151.9, CH | 151.9, CH | 150.6, CH |
| 16 | 108.4, CH2 | 112.7, CH2 | 112.7, CH2 | 110.4, CH2 | 110.4, CH2 | 112.9, CH2 | 112.7, CH2 | 110.5, CH2 | 110.4, CH2 | 109.6, CH2 |
| 17 | 28.5, CH3 | 29.2, CH3 | 29.1, CH3 | 22.6, CH3 | 22.6, CH3 | 26.8, CH3 | 29,2, CH3 | 22.6, CH3 | 22.6, CH3 | 21.7, CH3 |
| 18 | 25.8, CH3 | 22.8, CH3 | 23.7, CH3 | 23.9, CH3 | 24.6, CH3 | 23.6, CH3 | 23.7, CH3 | 22.0, CH3 | 24.6, CH3 | 23.9, CH3 |
| 19 | 74.0, CH2 | 73.3, CH2 | 73.8, CH2 | 73.9, CH2 | 74.1, CH2 | 73.1, CH2 | 73,6, CH2 | 75.1, CH2 | 73.9, CH2 | 72.1, CH2 |
| 20 | 17.7, CH3 | 18.7, CH3 | 19.7, CH3 | 16.9, CH3 | 17.6, CH3 | 16.1, CH3 | 19,7, CH3 | 16.7, CH3 | 17.6, CH3 | 15.7, CH3 |
| 1′ | 101.2, CH | 103.5, CH | 103.3, CH | 103.7, CH | 103.3, CH | 102.8, CH | 102.8, CH | 101.9, CH | 103.3, CH | 103.5, CH |
| 2′ | 72.7, CH | 71.1, CH | 71.3, CH | 70.9, CH | 71.2, CH | 71.6 CH | 71.7, CH | 72.1, CH | 71.3, CH | 71.6 CH |
| 3′ | 72.3, CH | 70.8, CH | 71.1, CH | 70.6, CH | 71.1, CH | 71.8, CH | 72.0, CH | 72.0, CH | 71.2, CH | 75.1, CH |
| 4′ | 67.2, CH | 69.9, CH | 69.7 CH | 69.9, CH | 69.6, CH | 69.1, CH | 69.0, CH | 68.7, CH | 69.7, CH | 70.0, CH |
| 5′ | 75.7, CH | 76.5, CH | 76.6, CH | 76.6, CH | 76.4, CH | 76.4, CH | 76.1, CH | 76.0, CH | 76.4, CH | 77.8, CH |
| 6′ | 64.0, CH2 | 174.0, C | 174.0, C | 172.7, C | 172.9, C | 173.2, C | 172.8, C | 172.6, C | 172.9, C | 170.7, C |
| 7′ | 53.3, CH3 | 53.3, CH3 | 53.4 CH3 | 53.3, CH3 | 53.3, CH3 | 66.8, CH2 | 51.8 CH3 | |||
| 8′ | 32.3, CH2 | |||||||||
| 9′ | 20.7, CH2 | |||||||||
| 10′ | 14.7, CH3 |
| Position | 1a | 2b | 3b | 4c | 5b |
|---|---|---|---|---|---|
| 1 | α: 1.54 t (11.5) β: 2.34 dd (4.6, 12.2) | α: 1.35 m β: 1.94 m | α: 1.23 m β: 2.17 dd (4.5, 12.8) | α: 1.22 dt (4.6, 13.5) β: 1.90 dd (3.5, 13.5) | α: 1.11 β: 2.11 dd (4.2, 12.6) |
| 2 | 4.28 ddd (4.5, 9.3, 11.5) | α: 1.82 dd (3.5, 11.9) β: 1.75 dd (4.0, 13.6) | 3.82 m | α: 1.74 dd (3.4, 11.8) β: 1.65 dd (3.0, 13.4) | 3.76 m |
| 3 | 3.61 d (9.3) | 3.26 dd (4.0, 11.9) | 2.99 d (9.8) | 3.24 dd (4.1, 11.8) | 2.98 d (9.8) |
| 5 | 2.41 d (13.2) | 1. 67 dd (3.6, 14.4) | 1. 76 dd (3.5, 14.7) | 1. 26 t (8.2) | 1.34 dd (3.9, 11.4) |
| 6 | 3.70 d (13.2) | α: 2.54 dd (3.6, 18.0) β: 2.64 dd (14.4, 18.0) | α: 2.56 dd (3.3, 18.2) β: 2.70 dd (14.7, 18.2) | 2.03 m | 2.03 m |
| 7 | 5.38 brs | 5.39 brs | |||
| 9 | 1.93 t (7.5) | 1.66 dd (3.9, 7.8) | 1.74 m | ||
| 11 | α: 1.38 m β: 1.69 m | α: 2.23 m β: 2.27 m | α: 2.26 m β: 2.32 m | α: 1.38 m β: 1.58 m | α: 1.41 m β: 1.61 m |
| 12 | α: 1.87 β: 1.36 | α: 1.35 m β: 1.62 m | α: 1.64 m β: 1.36 m | α: 1.37 m β: 1.48 dt (2.7, 8.9) | α: 1.50 dd (2.8, 12.1) β: 1.39 td (2.8, 11.5) |
| 14 | α: 2.37 d (14.0) β: 1.48 d (14.0) | α: 2.30 d (17.5) β: 1.93 d (17.5) | α: 2.31 m β: 1.94 dt (2.6, 17.9) | α: 1.97 brd (14.1) β: 1.91 dd (2.6, 14.1) | α: 1.99 m β: 1.92 dd (2.6, 14.1) |
| 15 | 6.64 dd (10.8, 17.6) | 5.70 dd (10.6, 17.5) | 5.70 dd (10.8, 17.5) | 5.80 dd (10.7, 17.5) | 5.81 dd (10.8, 17.6) |
| 16 | a: 4.85 dd (1.8, 10.8) b: 4.96 dd (1.8, 17.6) | a: 4.82 dd (1.4, 17.5) b: 4.93 dd (1.4, 10.6) | a: 4.82 dd (1.5, 17.6) b: 4.93 dd (1.5, 10.8) | a: 4.84 dd (1.3, 10.7) b: 4.92 dd (1.3, 17.5) | a: 4.85 dd (1.4, 10.8) b: 4.93 dd (1.4, 17.6) |
| 17 | 0.95 s | 1.00 s | 1.01 s | 0.86 s | 0.86 s |
| 18 | 1.81 s | 1.13 s | 1.16 s | 1.10 s | 1.11 s |
| 19 | a: 4.23 d (9.9) b: 4.98 d (9.9) | a: 3.73 d (10.2) b: 4.17 d (10.2) | a: 3.67 d (10.4) b: 4.14 d (10.4) | a: 3.83 d (10.2) b: 4.04 d (10.2) | a: 3.72 d (10.3) b: 4.03 d (10.3) |
| 20 | 1.28 s | 1.14 s | 1.21 s | 0.87 s | 0.95 s |
| 1 | 5.43 d (1.2) | 4.84 d (2.9) | 4.82 d (2.5) | 4.85 d (2.5) | 4.85 d (2.7) |
| 2′ | 4.56 dd (1.2, 3.9) | 3.77 dd (2.7, 7.8) | 3.77 dd (2.5, 7.4) | 3.77 dd (2.8, 7.9) | 3.77 m |
| 3′ | 4.74 t (3.7) | 3.93 dd (2.9, 7.7) | 3.94 dd (3.0, 7.4) | 3.89 dd (3.0, 7.9) | 3.90 dd (3.3, 7.3) |
| 4′ | 4.50 m | 4.23 t (4.8) | 4.20 dd (3.0, 5.7) | 4.26 dd (3.0, 4.9) | 4.23 dd (3.3, 5.6) |
| 5′ | 4.69 d (3.2, 12.2) | 4.24 d (4.8) | 4.22 d (5.7) | 4.28 d (4.9) | 4.28 d (5.6) |
| 6′ | a: 4.40 dd (6.5, 12.3) b: 4.51 m | ||||
| 7′ | 3.78 s | 3.78 s |
| Position | 6c | 7b | 8c | 9b | 10d |
|---|---|---|---|---|---|
| 1 | α: 1.22 m β: 2.07 dd (4.3, 12.7) | α: 1.23 m β: 2.18 dd (4.5, 12.8) | α: 1.50 m β: 2.19 m | α: 1.11 m β: 2.11 dd (4.2, 12.5) | α: 1.15 dt β: 1.78 brd (3.9, 13.1) |
| 2 | 3.79 m | 3.80 dd (9.8, 13.9) | α: 2.84 dt (5.4, 14.2) β: 2.23 m | 3.76 m | α: 1.85 m β: 1.92 m |
| 3 | 3.04 d (9.9) | 3.00 d (9.8) | 2.98 d (9.8) | 3.55 dd (4.0, 11.9) | |
| 5 | 1.73 dd (5.0, 13.8) | 1. 75 dd (3.4, 14.7) | 1.63 dd (4.1, 12.3) | 1.34 dd (4.5, 11.8) | 1.27 m |
| 6 | α: 2.59 dd (5.0, 19.0) | α: 2.53 dd (3.4, 18.2) β: 2.80 dd (14.7, 18.2) | α: 2.04 m β: 2.11 m | α: 2.01 m β: 2.06 m | α: 2.06 m β: 2.40 m |
| 7 | 5.41 brs | 5.38 m | 5.30 m | ||
| 9 | 2.13 m | 1.76 m | 1.74 m | 1.60 m | |
| 11 | α: 1.79 m β: 1.54 m | α: 2.26 m β: 2.31 m | α: 1.64 m β: 1.47 m | α: 1.41 m β: 1.61 m | α: 1.46 m β: 1.32 m |
| 12 | α: 1.54 m β: 1.67 m | α: 1.64 m β: 1.36 m | α: 1.51 β: 1.44 | α: 1.50 m β: 1.39 m | α: 1.32 m β: 1.45 m |
| 14 | 6.68 t (2.1) | α: 2.32 m β: 1.95 d (17.8) | α: 2.00 m β: 1.94 d (2.6, 14.0) | α: 1.99 m β: 1.92 dd (2.6, 14.1) | α: 2.03 brd (14.0) β: 1.94 brd (14.0) |
| 15 | 5.83 dd (10.7, 17.5) | 5.71 dd (10.8, 17.5) | 5.81 dd (10.7, 17.5) | 5.81 dd (10.8, 17.4) | 5.87 dd (10.6, 17.4) |
| 16 | 5.00 m | a: 4.83 dd (1.2, 17.5) b: 4.93 dd (1.2, 10.8) | a: 4.86 dd (1.4, 10.7) b: 4.94 dd (1.4, 17.5) | a: 4.85 dd (1.4, 10.8) b: 4.93 dd (1.4, 17.4) | a: 4.95 d (10.6) b: 5.02 d (17.4) |
| 17 | 1.12 s | 1,01 s | 0.89 s | 0.86 s | 0.90 s |
| 18 | 1.13 s | 1.15 s | 1.11 s | 1.11 s | 1.41 s |
| 19 | a: 3.68 d (10.3) b: 4.09 d (10.3) | a: 3.65 d (10,4) b: 4.11 d (10,4) | a: 3.90 d (9.8) b: 3.96 d (9.8) | a: 3.71 d (10.5) b: 4.04 d (1053) | a: 4.26 d (10.3) b: 4.59 d (10.3) |
| 20 | 0.95 s | 1.22 s | 1.17 s | 0.95 s | 0.93 s |
| 1′ | 4.78 brs | 4.79 d (2.0) | 4.75 d (1.9) | 4.84 d (2.1) | 4.97 brs |
| 2′ | 3.76 m | 3.76 m | 3.66 dd (1.9, 5.6) | 3.77 dd (2.5, 7.4) | 4.55 d (3.2) |
| 3′ | 3.91 dd (3.3, 6.3) | 3.92 dd (3.3, 6.0) | 3.89 dd (3.0, 5.6) | 3.92 dd (3.0, 7.4) | 4.14 dd (3.3, 9.4) |
| 4′ | 4.15 dd (3.3, 7.0) | 4.14 dd (3.3, 7.3) | 4.08 dd (3.2, 8.0) | 4.22 m | 4.87 t (9.3) |
| 5′ | 4.24 d (7.0) | 4.24 d (7.3) | 4.23 d (8.0) | 4.25 d (5.6) | 4.40 d (9.3) |
| 7′ | 3.76 s | 3.76 s | 3.77 s | a: 4.15 dt (6.6, 10.7) b: 4.19 m | 3.64 s |
| 8′ | a,b: 1.68 m | ||||
| 9′ | a,b: 1.45 m | ||||
| 10′ | 0.96 t (7.5) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuravleva, O.I.; Antonov, A.S.; Oleinikova, G.K.; Khudyakova, Y.V.; Popov, R.S.; Denisenko, V.A.; Pislyagin, E.A.; Chingizova, E.A.; Afiyatullov, S.S. Virescenosides from the Holothurian-Associated Fungus Acremonium striatisporum Kmm 4401. Mar. Drugs 2019, 17, 616. https://doi.org/10.3390/md17110616
Zhuravleva OI, Antonov AS, Oleinikova GK, Khudyakova YV, Popov RS, Denisenko VA, Pislyagin EA, Chingizova EA, Afiyatullov SS. Virescenosides from the Holothurian-Associated Fungus Acremonium striatisporum Kmm 4401. Marine Drugs. 2019; 17(11):616. https://doi.org/10.3390/md17110616
Chicago/Turabian StyleZhuravleva, Olesya I., Alexandr S. Antonov, Galina K. Oleinikova, Yuliya V. Khudyakova, Roman S. Popov, Vladimir A. Denisenko, Evgeny A. Pislyagin, Ekaterina A. Chingizova, and Shamil Sh. Afiyatullov. 2019. "Virescenosides from the Holothurian-Associated Fungus Acremonium striatisporum Kmm 4401" Marine Drugs 17, no. 11: 616. https://doi.org/10.3390/md17110616
APA StyleZhuravleva, O. I., Antonov, A. S., Oleinikova, G. K., Khudyakova, Y. V., Popov, R. S., Denisenko, V. A., Pislyagin, E. A., Chingizova, E. A., & Afiyatullov, S. S. (2019). Virescenosides from the Holothurian-Associated Fungus Acremonium striatisporum Kmm 4401. Marine Drugs, 17(11), 616. https://doi.org/10.3390/md17110616