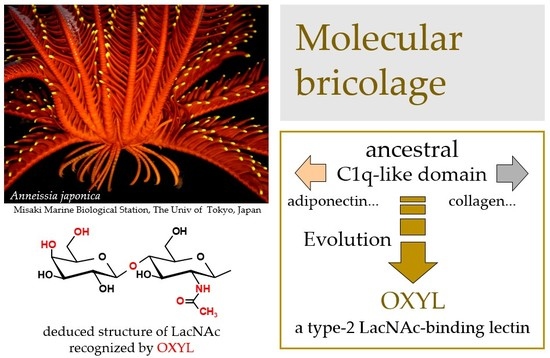
Functional Characterization of OXYL, A SghC1qDC LacNAc-specific Lectin from The Crinoid Feather Star Anneissia Japonica
Abstract
:1. Introduction
2. Results
2.1. Structural Characterization of OXYL as a sghC1qDC Protein
2.2. OXYL is Part of a Multigenic Family of sghC1qDC Proteins Restricted to Comatulida
2.3. Tetrameric Structure
2.4. Generation of Antiserum Against OXYL
2.5. Tissue Localization of OXYL
2.6. OXYL Quantitatively Binds to Type-2 N-LacNAc
2.7. OXYL Displays Bacteria Agglutination Properties
2.8. Antibiofilm Activity and Influence on Bacterial Growth of OXYL
2.9. OXYL Adhered to Cells by Binding to LacNAc on the Cell Surface
2.10. OXYL LacNAc-Dependently Activated the p38 of BT-474 Cancer Cells
3. Discussion
4. Experimental Design
4.1. Materials
4.2. Purification of the OXYL Lectin
4.3. Identification of the OXYL Coding Sequence
4.4. Phylogeny of OXYL
4.5. Molecular Mass Determination of OXYL
4.6. Generation of Antiserum and Evaluation of Anti-OXYL Antibody Specificity
4.7. Immunohistochemistry of OXYL in Feather Star Tissues
4.8. Sugar and Glycoconjugates-Binding Specificity of OXYL
4.9. Glycan Array Analysis
4.10. Bacteriostatic Assay
4.11. Anti-Biofilm Activity of OXYL
4.12. Binding of the Surface of Bacteria and Cultured Cancer Cells by OXYL
4.13. Detection of Activated Signal Transduction Molecules and their Phosphorylated Forms in BT-474 Cells in the Presence of OXYL
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vilanova, E.; Santos, G.R.; Aquino, R.S.; Valle-Delgado, J.J.; Anselmetti, D.; Fernàndez-Busquets, X.; Mourão, P.A. Carbohydrate-carbohydrate interactions mediated by sulfate esters and calcium provide the cell adhesion required for the emergence of early metazoans. J. Biol. Chem. 2016, 291, 9425–9437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karakostis, K.; Costa, C.; Zito, F.; Matranga, V. Heterologous expression of newly identified galectin-8 from sea urchin embryos produces recombinant protein with lactose binding specificity and anti-adhesive activity. Sci. Rep. 2015, 5, 17665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giga, Y.; Ikai, A.; Takahashi, K. The complete amino acid sequence of echinoidin, a lectin from the coelomic fluid of the sea urchin Anthocidaris crassispina. Homologies with mammalian and insect lectins. J. Biol. Chem. 1987, 262, 6197–6203. [Google Scholar] [PubMed]
- Flores, R.L.; Livingston, B.T. The skeletal proteome of the sea star Patiria miniata and evolution of biomineralization in echinoderms. BMC Evol. Biol. 2017, 17, 125. [Google Scholar] [CrossRef] [PubMed]
- Ozeki, Y.; Matsui, T.; Suzuki, M.; Titani, K. Amino acid sequence and molecular characterization of a D-galactoside-specific lectin purified from sea urchin (Anthocidaris crassispina) eggs. Biochemistry 1991, 30, 2391–2394. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, R.F.; Teixeira, C.S.; de Melo, A.A.; de Almeida, A.S.; Cavada, B.S.; de Sousa, O.V.; da Rocha, B.A.; Nagano, C.S.; Sampaio, A.H. L-Rhamnose-binding lectin from eggs of the Echinometra lucunter: Amino acid sequence and molecular modeling. Int. J. Biol. Macromol. 2015, 78, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Uchida, T.; Yamasaki, T.; Eto, S.; Sugawara, H.; Kurisu, G.; Nakagawa, A.; Kusunoki, M.; Hatakeyama, T. Crystal structure of the hemolytic lectin CEL-III isolated from the marine invertebrate Cucumaria echinata: implications of domain structure for its membrane pore-formation mechanism. J. Biol. Chem. 2004, 279, 37133–37141. [Google Scholar] [CrossRef] [PubMed]
- Maehashi, E.; Sato, C.; Ohta, K.; Harada, Y.; Matsuda, T.; Hirohashi, N.; Lennarz, W.J.; Kitajima, K. Identification of the sea urchin 350-kDa sperm-binding protein as a new sialic acid-binding lectin that belongs to the heat shock protein 110 family: implication of its binding to gangliosides in sperm lipid rafts in fertilization. J. Biol. Chem. 2003, 278, 42050–42057. [Google Scholar] [CrossRef] [PubMed]
- Pohleven, J.; Renko, M.; Magister, Š.; Smith, D.F.; Künzler, M.; Štrukelj, B.; Turk, D.; Kos, J.; Sabotiè, J. Bivalent carbohydrate binding is required for biological activity of Clitocybe nebularis lectin (CNL), the N,Nʹ-diacetyllactosediamine (GalNAcβ1-4GlcNAc, LacdiNAc)-specific lectin from basidiomycete C. nebularis. J. Biol. Chem. 2012, 287, 10602–10612. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Fujiwara, T.; Koide, Y.; Hasan, I.; Sugawara, S.; Rajia, S.; Kawsar, S.M.; Yamamoto, D.; Araki, D.; Kanaly, R.A.; et al. Internalization of a novel, huge lectin from Ibacus novemdentatus (slipper lobster) induces apoptosis of mammalian cancer cells. Glycoconj. J. 2017, 34, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, E.V.M.M.; Oliveira, W.F.; Coelho, L.C.B.B.; Correia, M.T.S. Lectins as mitosis stimulating factors: Briefly reviewed. Life Sci. 2018, 207, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Hasan, I.; Ozeki, Y. Histochemical localization of N-acetylhexosamine-binding lectin HOL-18 in Halichondria okadai (Japanese black sponge), and its antimicrobial and cytotoxic anticancer effects. Int. J. Biol. Macromol. 2019, 124, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Ozeki, Y.; Matsui, T.; Titani, K. Cell adhesive activity of two animal lectins through different recognition mechanisms. FEBS Lett. 1991, 289, 145–147. [Google Scholar] [CrossRef] [Green Version]
- Hatakeyama, T.; Ichise, A.; Unno, H.; Goda, S.; Oda, T.; Tateno, H.; Hirabayashi, J.; Sakai, H.; Nakagawa, H. Carbohydrate recognition by the rhamnose-binding lectin SUL-I with a novel three-domain structure isolated from the venom of globiferous pedicellariae of the flower sea urchin Toxopneustes pileolus. Protein Sci. 2017, 26, 1574–1583. [Google Scholar] [CrossRef] [PubMed]
- Zamora, S.; Rahman, I.A.; Ausich, W.I. Palaeogeographic implications of a new iocrinid crinoid (Disparida) from the Ordovician (Darriwillian) of Morocco. PeerJ. 2015, 3, e1450. [Google Scholar] [CrossRef] [PubMed]
- Oji, T.; Twitchett, R.J. The oldest post-Palaeozoic Crinoid and Permian-Triassic origins of the Articulata (Echinodermata). Zoolog. Sci. 2015, 32, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Kondo, M.; Akasaka, K. Regeneration in crinoids. Dev. Growth Differ. 2010, 52, 57–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibata, T.F.; Sato, A.; Oji, T.; Akasaka, K. Development and growth of the feather star Oxycomanthus japonicus to sexual maturity. Zoolog. Sci. 2008, 25, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Summers, M.M.; Messing, C.G.; Rouse, G.W. Phylogeny of Comatulidae (Echinodermata: Crinoidea: Comatulida): a new classification and an assessment of morphological characters for crinoid taxonomy. Mol. Phylogenet. Evol. 2014, 80, 319–339. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, R.; Shibata, T.F.; Kohtsuka, H.; Sekifuji, M.; Sugii, N.; Nakajima, H.; Kojima, N.; Fujii, Y.; Kawsar, S.M.; Yasumitsu, H.; et al. Glycomics of a novel type-2 N-acetyllactosamine-specific lectin purified from the feather star, Oxycomanthus japonicus (Pelmatozoa: Crinoidea). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 158, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Sequencing The Genome of An Early Branching Echinoderm; The Crinoid Oxycomanthus Japonicus. Available online: https://www.ncbi.nlm.nih.gov/bioproject/236227 (accessed on 24 February 2019).
- Takagaki, Y.; Manley, J.L. R.N.A. recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 1997, 17, 3907–3914. [Google Scholar] [CrossRef] [PubMed]
- Ghai, R.; Waters, P.; Roumenina, L.; Gadjeva, M.; Kojouharova, M.S.; Reid, K.B.; Sim, R.B.; Kishore, U. C1q and its growing family. Immunobiology 2007, 212, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Carland, T.M.; Gerwick, L. The C1q domain containing proteins: Where do they come from and what do they do? Dev. Comp. Immunol. 2010, 34, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; Venier, P.; Pallavicini, A. The genome of the Pacific oyster Crassostrea gigas brings new insights on the massive expansion of the C1q gene family in Bivalvia. Dev. Comp. Immunol. 2015, 49, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Ozeki, Y.; Yokota, Y.; Kato, K.H.; Titani, K.; Matsui, T. Developmental expression of D-galactoside-binding lectin in sea urchin (Anthocidaris crassispina) eggs. Exp. Cell Res. 1995, 216, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Alliegro, M.C.; Alliegro, M.A. Echinonectin is a Del-1-like molecule with regulated expression in sea urchin embryos. Gene Expr. Patterns 2007, 7, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Hasan, I.; Sugawara, S.; Fujii, Y.; Koide, Y.; Terada, D.; Iimura, N.; Fujiwara, T.; Takahashi, K.G.; Koima, N.; Rajia, S.; et al. MytiLec, a mussel R-type lectin, interacts with surface glycan Gb3 on Burkitt’s lymphoma cells to trigger apoptosis through multiple pathways. Mar. Drugs. 2015, 13, 7377–7389. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, S.; Im, C.; Kawano, T.; Tatsuta, T.; Koide, Y.; Yamamoto, D.; Ozeki, Y.; Nitta, K.; Hosono, M. Catfish rhamnose-binding lectin induces G(0/1) cell cycle arrest in Burkitt’s lymphoma cells via membrane surface Gb3. Glycoconj. J. 2017, 34, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; Manfrin, C.; De Moro, G.; Figueras, A.; Novoa, B.; Venier, P.; Pallavicini, A. The C1q domain containing proteins of the Mediterranean mussel Mytilus galloprovincialis: a widespread and diverse family of immune-related molecules. Dev. Comp. Immunol. 2011, 35, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Allam, B.; Pales Espinosa, E.; Tanguy, A.; Jeffroy, F.; Le Bris, C.; Paillard, C. Transcriptional changes in Manila clam (Ruditapes philippinarum) in response to Brown Ring Disease. Fish Shellfish Immunol. 2014, 41, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H.; Xiang, L.X.; Shao, J.Z. A novel C1q-domain-containing (C1qDC) protein from Mytilus coruscus with the transcriptional analysis against marine pathogens and heavy metals. Dev. Comp. Immunol. 2014, 44, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Tills, O.; Truebano, M.; Feldmeyer, B.; Pfenninger, M.; Morgenroth, H.; Schell, T.; Rundle, S.D. Transcriptomic responses to predator kairomones in embryos of the aquatic snail Radix balthica. Ecol. Evol. 2018, 8, 11071–11082. [Google Scholar] [CrossRef] [PubMed]
- Gross, P.S.; Al-Sharif, W.Z.; Clow, L.A.; Smith, L.C. Echinoderm immunity and the evolution of the complement system. Dev. Comp. Immunol. 1999, 23, 429–442. [Google Scholar] [CrossRef]
- Buckley, K.M.; Smith, L.C. Extraordinary diversity among members of the large gene family, 185/333, from the purple sea urchin, Strongylocentrotus purpuratus. BMC Mol. Biol. 2007, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Ressl, S.; Vu, B.K.; Vivona, S.; Martinelli, D.C.; Südhof, T.C.; Brunger, A.T. Structures of C1q-like proteins reveal unique features among the C1q/TNF superfamily. Structure. 2015, 23, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Kishore, U.; Gaboriaud, C.; Waters, P.; Shrive, A.K.; Greenhough, T.J.; Reid, K.B.; Sim, R.B.; Arlaud, G.J. C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol. 2004, 25, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, L.; Scherer, P.E. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 1998, 8, 335–338. [Google Scholar] [CrossRef]
- Surolia, A.; Swaminathan, C.P.; Ramkumar, R.; Podder, S.K. Unusual structural stability and ligand induced alterations in oligomerization of a galectin. FEBS Lett. 1997, 409, 417–420. [Google Scholar] [CrossRef] [Green Version]
- Sulák, O.; Cioci, G.; Delia, M.; Lahmann, M.; Varrot, A.; Imberty, A.; Wimmerová, M.A. TNF-like trimeric lectin domain from Burkholderia cenocepacia with specificity for fucosylated human histo-blood group antigens. Structure 2010, 18, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yu, S.; Zhao, J.; Su, X.; Li, T. Cloning and characterization of a sialic acid binding lectins (SABL) from Manila clam Venerupis philippinarum. Fish Shellfish Immunol. 2011, 30, 1202–1206. [Google Scholar] [CrossRef] [PubMed]
- Kong, P.; Zhang, H.; Wang, L.; Zhou, Z.; Yang, J.; Zhang, Y.; Qiu, L.; Wang, L.; Song, L. AiC1qDC-1, a novel gC1q-domain-containing protein from bay scallop Argopecten irradians with fungi agglutinating activity. Dev. Comp. Immunol. 2010, 34, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Qiu, L.; Wang, M.; Jia, Z.; Wang, W.; Xin, L.; Liu, Z.; Wang, L.; Song, L. Comparative study of three C1q domain containing proteins from pacific oyster Crassostrea gigas. Dev. Comp. Immunol. 2018, 78, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, O.; Wada, Y.; Namai, F.; Saito, E.; Araki, K.; Yamamoto, A.; Tsutsui, S. A novel C1q family member with fucose-binding activity from surfperch, Neoditrema ransonnetii (Perciformes, Embiotocidae). Fish Shellfish Immunol. 2009, 27, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M.; Matsushita, A.; Endo, Y.; Nakata, M.; Kojima, N.; Mizuochi, T.; Fujita, T. Origin of the classical complement pathway: Lamprey orthologue of mammalian C1q acts as a lectin. Proc. Natl. Acad. Sci. USA 2004, 101, 10127–10131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peschke, B.; Keller, C.W.; Weber, P.; Quast, I.; Lünemann, J.D. Fc-galactosylation of human immunoglobulin gamma isotypes improves C1q binding and enhances. Complement-dependent cytotoxicity. Front. Immunol 2017, 8, 646. [Google Scholar] [CrossRef] [PubMed]
- Païdassi, H.; Tacnet-Delorme, P.; Lunardi, T.; Arlaud, G.J.; Thielens, N.M.; Frachet, P. The lectin-like activity of human C1q and its implication in DNA and apoptotic cell recognition. FEBS Lett. 2008, 582, 3111–3116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garlatti, V.; Chouquet, A.; Lunardi, T.; Vivès, R.; Païdassi, H.; Lortat-Jacob, H.; Thielens, N.M.; Arlaud, G.J.; Gaboriaud, C. Cutting edge: C1q binds deoxyribose and heparan sulfate through neighboring sites of its recognition domain. J. Immunol. 2010, 185, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Peake, P.W.; Shen, Y.; Campbell, L.V.; Charlesworth, J.A. Human adiponectin binds to bacterial lipopolysaccharide. Biochem. Biophys. Res. Commun. 2006, 341, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Gaboriaud, C.; Juanhuix, J.; Gruez, A.; Lacroix, M.; Darnault, C.; Pignol, D.; . Verger, D.; Fontecilla-Camps, J.C.; Arlaud, G.J. The Crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J. Biol. Chem. 2003, 278, 46974–46982. [Google Scholar] [CrossRef] [PubMed]
- Pajvani, U.B.; Du, X.; Combs, T.P.; Berg, A.H.; Rajala, M.W.; Schulthess, T.; Engel, J.; Brownlee, M.; Scherer, P.E. Structure-function studies of the adipocyte-secreted hormone Acrp30/adiponectin. Implications fpr metabolic regulation and bioactivity. J. Biol. Chem. 2003, 278, 9073–9085. [Google Scholar] [CrossRef] [PubMed]
- Bao, D.; Pang, Z.; Morgan, J.I. The structure and proteolytic processing of Cbln1 complexes. J. Neurochem. 2005, 95, 618–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsao, T.-S.; Murrey, H.E.; Hug, C.; Lee, D.H.; Lodish, H.F. Oligomerization state-dependent activation of NF-kappa B signaling pathway by adipocyte complement-related protein of 30 kDa (Acrp30). J. Biol. Chem. 2003, 277, 29359–29362. [Google Scholar] [CrossRef] [PubMed]
- Tsao, T.-S.; Tomas, E.; Murrey, H.E.; Hug, C.; Lee, D.H.; Ruderman, N.B.; Heuser, J.E.; Lodish, H.F. Role of disulfide bonds in Acrp30/adiponectin structure and signaling specificity. Different oligomers activate different signal transduction pathways. J. Biol. Chem. 2003, 278, 50810–50817. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman Girija, U.; Gingras, A.R.; Marshall, J.E.; Panchal, R.; Sheikh, M.A.; Harper, J.A.J.; Gál, P.; Schwaeble, W.J.; Mitchell, D.A.; Moody, P.C.E.; et al. Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation. Proc. Natl. Acad. Sci. USA 2013, 110, 13916–13920. [Google Scholar] [CrossRef] [PubMed]
- Agostinis, C.; Vidergar, R.; Belmonte, B.; Mangogna, A.; Amadio, L.; Geri, P.; Borelli, V.; Zanconati, F.; Tedesco, F.; Confalonieri, M.; et al. Complement protein C1q binds to hyaluronic acid in the malignant pleural mesothelioma microenvironment and promotes tumor growth. Front. Immunol. 2017, 8, 1559. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Xiang, J.; Lu, Y.; Chen, Y.; Wang, T.; Gong, G.; Wang, L.; Li, X.; Chen, S.; Sha, Z. sghC1q, a novel C1q family member from half-smooth tongue sole (Cynoglossus semilaevis): identification, expression and analysis of antibacterial and antiviral activities. Dev. Comp. Immunol. 2015, 48, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fan, C.; Xu, W.; Zhang, Y.; Dong, Z.; Xiang, J.; Chen, S. Characterization and functional analysis of a novel C1q-domain-containing protein in Japanese flounder (Paralichthys olivaceus). Dev Comp. Immunol. 2017, 67, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Bulla, R.; Tripodo, C.; Rami, D.; Ling, G.S.; Agostinis, C.; Guarnotta, C.; Zorzet, S.; Durigutto, P.; Botto, M.; Tedesco, F. C1q acts in the tumour microenvironment as a cancer-promoting factor independently of complement activation. Nat. Commun. 2016, 7, 10346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pier, G.B. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int. J. Med. Microbiol. 2007, 297, 277–295. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, R.F.; Torres, R.C.; Chaves, R.P.; de Vasconcelos, M.A.; de Sousa, B.L.; Goveia, A.C.; Arruda, F.V.; Matos, M.N.; Matthews-Cascon, H.; Freire, V.N.; et al. Purification, biochemical characterization, and amino acid sequence of a novel type of lectin from Aplysia dactylomela eggs with antibacterial/antibiofilm potential. Mar. Biotechnol. (NY) 2017, 19, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.C.; Fabres-Klein, M.H.; de Oliveira, L.L.; Feio, R.N.; Malouin, F.; Ribon Ade, O.A. C-type lectin from Bothrops jararacussu venom disrupts Staphylococcal biofilms. PLoS One 2015, 10, e0120514. [Google Scholar] [CrossRef] [PubMed]
- Terada, D.; Voet, A.R.D.; Noguchi, H.; Kamata, K.; Ohki, M.; Addy, C.; Fujii, Y.; Yamamoto, D.; Ozeki, Y.; Tame, J.R.H.; et al. Computational design of a symmetrical β-trefoil lectin with cancer cell binding activity. Sci. Rep. 2017, 7, 5943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, Y.; Sugawara, S.; Araki, D.; Kawano, T.; Tatsuta, T.; Takahashi, K.; Kawsar, S.M.; Matsumoto, R.; Kanaly, R.A.; Yasumitsu, H.; et al. MRP1 expressed on Burkitt’s lymphoma cells was depleted by catfish egg lectin through Gb3-glycosphingolipid and enhanced cytotoxic effect of drugs. Protein J. 2012, 31, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Wada, J.; Ota, K.; Kumar, A.; Wallner, E.I.; Kanwar, Y.S. Developmental regulation, expression, and apoptotic potential of galectin-9, a beta-galactoside binding lectin. J. Clin. Invest. 1997, 99, 2452–2461. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, J.; Hashidate, T.; Arata, Y.; Nishi, N.; Nakamura, T.; Hirashima, M.; Urashima, T.; Oka, T.; Futai, M.; Muller, W.E.; et al. Oligosaccharide specificity of galectins: a search by frontal affinity chromatography. Biochim. Biophys. Acta 2002, 1572, 232–254. [Google Scholar] [CrossRef]
- Iwaki, J.; Hirabayashi, J. Carbohydrate-binding specificity of human galecins: An overview by frontal affinity chromatography. Trends Glycosci. Glycotechnol 2018, 30, 137–153. [Google Scholar] [CrossRef]
- Canesi, L.; Ciacci, C.; Fabbri, R.; Balbi, T.; Salis, A.; Damonte, G.; Cortese, K.; Caratto, V.; Monopoli, M.P.; Dawson, K.; et al. Interactions of cationic polystyrene nanoparticles with marine bivalve hemocytes in a physiological environment: Role of soluble hemolymph proteins. Environ. Res. 2016, 150, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Schmid, A.; Kopp, A.; Hanses, F.; Karrasch, T.; Schäffler, A. C1q/TNF-related protein-3 (CTRP-3) attenuates lipopolysaccharide (LPS)-induced systemic inflammation and adipose tissue Erk-1/-2 phosphorylation in mice in vivo. Biochem. Biophys. Res. Commun. 2014, 452, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M. The complement system of agnathans. Front. Immunol. 2018, 18, 1405. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Jiang, S.; Wang, M.; Wang, L.; Chen, H.; Xu, J.; Lv, Z.; Song, L. A novel siglec (CgSiglec-1) from the Pacific oyster (Crassostrea gigas) with broad recognition spectrum and inhibitory activity to apoptosis, phagocytosis and cytokine release. Dev. Comp. Immunol. 2016, 61, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Pallen, M.J.; Gophna, U. Bacterial flagella and Type III secretion: case studies in the evolution of complexity. Genome Dyn. 2007, 3, 30–47. [Google Scholar] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Krohn, R.I.; Gartner, F.T.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Wiechelman, K.J.; Braun, R.D.; Fitzpatrick, J.D. Investigation of the bicinchoninic acid protein assay: Identification of the groups responsible for color formation. Anal. Biochem. 1988, 75, 231–237. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, N.J. Ending the message: poly(A) signals then and now. Genes Dev. 2011, 25, 1770–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinforma. Oxf. Engl. 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: selection of best-fit models of protein evolution. Bioinforma. Oxf. Engl. 2005, 21, 2104–2105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuck, P. Sedimentation analysis of noninteracting and self-associating solutes using numerical solutions to the Lamm equation. Biophys. J. 1998, 75, 1503–1512. [Google Scholar] [CrossRef]
- Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000, 78, 1606–1619. [Google Scholar] [CrossRef]
- Goldman, A.; Ursitti, J.A.; Mozdzanowski, J.; Speicher, D.W. Electroblotting from polyacrylamide gels. Curr. Protoc. Protein. Sci. 2015, 82, 1–16. [Google Scholar]
- Shibata, T.F.; Oji, T.; Akasaka, K.; Agata, K. Staging of regeneration process of an arm of the feather star Oxycomanthus japonicus focusing on the oral-aboral boundary. Dev. Dyn. 2010, 239, 2947–2961. [Google Scholar] [CrossRef] [PubMed]
- Gourdine, J.P.; Cioci, G.; Miguet, L.; Unverzagt, C.; Silva, D.V.; Varrot, A.; Gautier, C.; Smith-Ravin, E.J.; Imberty, A. High affinity interaction between a bivalve C-type lectin and a biantennary complex-type N-glycan revealed by crystallography and microcalorimetry. J. Biol. Chem. 2008, 283, 30112–30120. [Google Scholar] [CrossRef] [PubMed]










| No. Name | Structures |
|---|---|
| 1. Lac | Galβ1-4Glc |
| 2. S3LN | NeuAcα2-3Galβ1-4GlcNAc |
| 3. S6LN | NeuAcα2-6Galβ1-4GlcNAc |
| 4. LNnT | Galβ1-3GlcNAcβ1-3Galβ1-4Glc |
| 5. LNT | Galβ1-3GlcNAcβ1-3Galβ1-4Glc |
| 6. NA2 |  |
| 7. NA3 |  |
| 8. NA4 |  |
| Saccharides | Minimum inhibitory conc. (mM) |
| N-acetyllactosamine (Galβ1-4GlcNAc) | 3.13 |
| lactose (Galβ1-4Glc) | >100 2,3 |
| D-galactose (Gal) | >100 2,3 |
| N-acetylneuramic acid (NeuAc) | >100 2 |
| Glycosamino glycans (GAG) | Minimum inhibitory conc. (mg/mL) |
| chondroitin sulphate | >50 2 |
| heparin sodium | >50 2 |
| Lipopolysaccharide (LPS) and Peptidoglycan (PG) | Minimum inhibitory conc. (mg/mL) |
| P. aeruginosa lipopolysaccharide | >50 2 |
| M. luteus peptidoglycan | >50 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, I.; Gerdol, M.; Fujii, Y.; Ozeki, Y. Functional Characterization of OXYL, A SghC1qDC LacNAc-specific Lectin from The Crinoid Feather Star Anneissia Japonica. Mar. Drugs 2019, 17, 136. https://doi.org/10.3390/md17020136
Hasan I, Gerdol M, Fujii Y, Ozeki Y. Functional Characterization of OXYL, A SghC1qDC LacNAc-specific Lectin from The Crinoid Feather Star Anneissia Japonica. Marine Drugs. 2019; 17(2):136. https://doi.org/10.3390/md17020136
Chicago/Turabian StyleHasan, Imtiaj, Marco Gerdol, Yuki Fujii, and Yasuhiro Ozeki. 2019. "Functional Characterization of OXYL, A SghC1qDC LacNAc-specific Lectin from The Crinoid Feather Star Anneissia Japonica" Marine Drugs 17, no. 2: 136. https://doi.org/10.3390/md17020136
APA StyleHasan, I., Gerdol, M., Fujii, Y., & Ozeki, Y. (2019). Functional Characterization of OXYL, A SghC1qDC LacNAc-specific Lectin from The Crinoid Feather Star Anneissia Japonica. Marine Drugs, 17(2), 136. https://doi.org/10.3390/md17020136