Penicamide A, A Unique N,N′-Ketal Quinazolinone Alkaloid from Ascidian-Derived Fungus Penicillium sp. 4829
Abstract
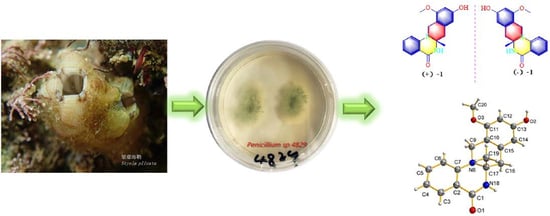
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Biological Material
3.3. Extraction, Isolation, and Characterization
3.3.1. Penicamide A (1)
- (+)-(1): + 28.5 (c 0.02, MeOH); ECD (MeOH) λmax (Δε): 209 (−1.8), 228 (+6.3), 257 (+5.6) nm.
- (−)-(1): − 30.1 (c 0.02, MeOH); ECD (MeOH) λmax (Δε): 210 (+1.0), 227 (−7.7), 257 (−6.1) nm.
3.3.2. Penicamide B (2)
3.4. X-ray Crystallographic Analysis of Compound 1
3.5. Calculation of the ECD Spectra
3.6. Cytotoxic Assay
3.7. Anti-Inflammation Bioassays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Palanisamy, S.K.; Rajendran, N.M.; Marino, A. Natural products diversity of marine ascidians (tunicates; ascidiacea) and successful drugs in clinical development. Nat. Prod. Bioprospect. 2017, 7, 1–111. [Google Scholar] [CrossRef]
- Watters, D.J. Ascidian toxins with potential for drug development. Mar. Drugs 2018, 16, 162. [Google Scholar] [CrossRef]
- von Schwarzenberg, K.; Vollmar, A.M. Targeting apoptosis pathways by natural compounds in cancer: Marine compounds as lead structures and chemical tools for cancer therapy. Cancer Lett. 2013, 332, 295–303. [Google Scholar] [CrossRef]
- D’Incalci, M.; Galmarini, C.M. A review of trabectedin (ET-743): A unique mechanism of action. Mol. Cancer Ther. 2010, 9, 2157–2163. [Google Scholar] [CrossRef]
- Schofield, M.M.; Jain, S.; Porat, D.; Gregory, J.D.; David, H.S. Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743. Environ. Microbiol. 2015, 17, 3964–3975. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Hu, J.S.; Xu, J.L.; Shao, C.L.; Wang, G.Y. Biological and chemical diversity of ascidian-associated microorganisms. Mar. Drugs 2018, 16, 362. [Google Scholar] [CrossRef]
- He, H.; Ding, W.-D.; Bernan, V.S.; Richardson, A.D.; Ireland, C.M.; Greenstein, M.; Ellestad, G.A.; Carter, G.T. Lomaiviticins A and B, potent antitumor antibiotics from micromonospora lomaivitiensis. J. Am. Chem. Soc. 2001, 123, 5362–5363. [Google Scholar] [CrossRef]
- Lin, Z.; Koch, M.; Abdel Aziz, M.H.; Galindo-Murillo, R.; Tianero, M.D.; Cheatham, T.E.; Barrows, L.R.; Reilly, C.A.; Schmidt, E.W. Oxazinin A, a pseudodimeric natural product of mixed biosynthetic origin from a filamentous fungus. Org. Lett. 2014, 16, 4774–4777. [Google Scholar] [CrossRef]
- Chen, S.; Shen, H.; Zhang, P.; Cheng, H.; Dai, X.; Liu, L. Anti-glioma trichobamide A with an unprecedented tetrahydro-5H-furo [2,3-b]pyrrol-5-one functionality from ascidian-derived fungus Trichobotrys effuse 4729. Chem. Commun. 2019, 55, 1438–1441. [Google Scholar] [CrossRef]
- Niaz, S.I.; Zhang, P.; Shen, H.; Li, J.; Chen, B.; Chen, S.; Liu, L.; He, J. Two new isochromane derivatives penisochromanes A and B from ascidian-derived fungus Penicillium sp. 4829. Nat. Prod. Res. 2019, 33, 1262–1268. [Google Scholar] [CrossRef]
- Zhuang, P.; Tang, X.-X.; Yi, Z.-W.; Qiu, Y.-K.; Wu, Z. Two new compounds from marine-derived fungus Penicillium sp. F11. J. Asian Nat. Prod. Res. 2012, 14, 197–203. [Google Scholar] [CrossRef]
- Shang, Z.; Li, X.; Meng, L.; Li, C.; Gao, S.; Huang, C.; Wang, B. Chemical profile of the secondary metabolites produced by a deep-sea sediment-derived fungus Penicillium commune SD-118. Chin. J. Oceanol. Limnol. 2012, 30, 305–314. [Google Scholar] [CrossRef]
- Bloch, P.; Tamm, C.; Bollinger, P.; Petcher, T.J.; Weber, H.P. Pseurotin, a new metabolite of Pseudeurotium ovalis STOLK having an unusual hetero-spirocyclic system. Helv. Chim. Acta 1976, 59, 133–137. [Google Scholar] [CrossRef]
- Wu, J.; Chen, X.; Su, J.; Lu, H.; Liu, Z. Chemical constituents of Bangia fuscopurpurea. Chem. Nat. Compd. 2019. [Google Scholar] [CrossRef]
- Özkaya, F.C.; Ebrahim, W.; Klopotowski, M.; Liu, Z.; Janiak, C.; Proksch, P. Isolation and X-ray structure analysis of citreohybridonol from marine-derived Penicillium atrovenetum. Nat. Prod. Res. 2018, 32, 840–843. [Google Scholar] [CrossRef]
- Kosemura, S.; Matsuo, S.; Yamamura, S. Citreohybriddione C, a meroterpenoid of a hybrid strain KO 0031 derived from Penicillium citreoviride B. IFO 6200 and 4692. Phytochemistry 1996, 43, 1231–1234. [Google Scholar] [CrossRef]
- Eguchi, S. Quinazoline alkaloids and related chemistry. In Bioactive Heterocycles I; Springer: Berlin/Heidelberg, Germany, 2006; pp. 113–156. [Google Scholar]
- Kshirsagar, U.A. Recent developments in the chemistry of quinazolinone alkaloids. Org. Biomol. Chem. 2015, 13, 9336–9352. [Google Scholar] [CrossRef]
- Shang, X.F.; Morris-Natschke, S.L.; Liu, Y.Q.; Guo, X.; Xu, X.S.; Goto, M.; Li, J.C.; Yang, G.Z.; Lee, K.H. Biologically active quinoline and quinazoline alkaloids part I. Med. Res. Rev. 2018, 38, 775–828. [Google Scholar] [CrossRef]
- Zheng, C.-J.; Li, L.; Zou, J.-P.; Han, T.; Qin, L.-P. Identification of a quinazoline alkaloid produced by Penicillium vinaceum, an endophytic fungus from Crocus sativus. Pharm. Biol. 2012, 50, 129–133. [Google Scholar] [CrossRef]
- Hwang, J.-M.; Oh, T.; Kaneko, T.; Upton, A.M.; Franzblau, S.G.; Ma, Z.; Cho, S.-N.; Kim, P. Design, synthesis, and structure–activity telationship studies of tryptanthrins as antitubercular agents. J. Nat. Prod. 2013, 76, 354–367. [Google Scholar] [CrossRef]
- Wang, X.; Yin, J.; Shi, L.; Zhang, G.; Song, B. Design, synthesis, and antibacterial activity of novel Schiff base derivatives of quinazolin-4(3H)-one. Eur. J. Med. Chem. 2014, 77, 65–74. [Google Scholar] [CrossRef]
- Yu, G.; Zhou, G.; Zhu, M.; Wang, W.; Zhu, T.; Gu, Q.; Li, D. Neosartoryadins A and B, fumiquinazoline alkaloids from a mangrove-derived fungus Neosartorya udagawae HDN13-313. Org. Lett. 2016, 18, 244–247. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Azuma, K.; Takakura, K.; Abe, H.; Kim, H.-S.; Wataya, Y.; Harayama, T. Asymmetric synthesis of (+)-febrifugine and (+)-isofebrifugine using yeast reduction. Tetrahedron 2001, 57, 1213–1218. [Google Scholar] [CrossRef]
- Molina, P.; Tárraga, A.; Gonzalez-Tejero, A.; Rioja, I.; Ubeda, A.; Terencio, M.C.; Alcaraz, M.J. Inhibition of leukocyte functions by the alkaloid isaindigotone from Isatis indigotica and some new synthetic derivatives. J. Nat. Prod. 2001, 64, 1297–1300. [Google Scholar] [CrossRef]
- Ma, Z.; Hano, Y.; Nomura, T.; Chen, Y.J.B. Novel quinazoline–quinoline alkaloids with cytotoxic and DNA topoisomerase II inhibitory activities. Bioorg. Med. Chem. Lett. 2004, 14, 1193–1196. [Google Scholar] [CrossRef]
- Kjer, J.; Debbab, A.; Aly, A.H.; Proksch, P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010, 5, 479. [Google Scholar] [CrossRef]
- Chen, S.; Ding, M.; Liu, W.; Huang, X.; Liu, Z.; Lu, Y.; Liu, H.; She, Z. Anti-inflammatory meroterpenoids from the mangrove endophytic fungus Talaromyces amestolkiae YX1. Phytochemistry 2018, 146, 8–15. [Google Scholar] [CrossRef]
- Chen, S.; Chen, D.; Cai, R.; Cui, H.; Long, Y.; Lu, Y.; Li, C.; She, Z. Cytotoxic and Antibacterial Preussomerins from the Mangrove Endophytic Fungus Lasiodiplodia theobromae ZJ-HQ1. J. Nat. Prod. 2016, 79, 2397–2402. [Google Scholar] [CrossRef]
- Zhang, P.; Deng, Y.; Lin, X.; Chen, B.; Li, J.; Liu, H.; Chen, S.; Liu, L. Anti-inflammatory Mono- and Dimeric Sorbicillinoids from the Marine-Derived Fungus Trichoderma reesei 4670. J. Nat. Prod. 2019, 82, 947–957. [Google Scholar] [CrossRef]






| No. | 1 | 2 | ||
|---|---|---|---|---|
| δC, Type | δH, (J in Hz) | δC, Type | δH, (J in Hz) | |
| 1 | 161.8, C | 169.4, C | ||
| 2 | 116.3, C | 117.8, C | ||
| 3 | 127.4, CH | 7.75, dd (1.6, 7.7) | 131.2, CH | 7.99, d (8.3) |
| 4 | 117.9, CH | 6.85, m | 123.5, CH | 7.19, t (7.4) |
| 5 | 133.9, CH | 7.45, t (8.5) | 133.8, CH | 7.62, m |
| 6 | 112.5, CH | 6.85, m | 127.4, CH | 8.47, d (8.3) |
| 7 | 147.1, C | 140.0, C | ||
| 8 | ||||
| 9 | 42.1, CH2 | 3.76, d (16.4) 4.45, d (16.4) | 163.0, C | |
| 10 | 109.9, C | 122.4, C | ||
| 11 | 156.6, C | 129.5, CH | 7.76, d (8.7) | |
| 12 | 97.1, CH | 6.32, d (1.6) | 115.9, CH | 6.95, d (8.7) |
| 13 | 157.2, C | 161.9, C | ||
| 14 | 106.3, CH | 6.16, d (1.6) | 115.9, CH | 6.95, d (8.7) |
| 15 | 132.8, C | 129.5, CH | 7.76, d (8.7) | |
| 16 | 41.7, CH2 | 2.80, d (15.9) 3.08, d (15.9) | 12.09, d (10.6) | |
| 17 | 68.9, C | 137.4, CH | 7.64, m | |
| 18 | 8.42, s | 100.2, CH | 5.46, d (8.7) | |
| 19 | 21.3, CH3 | 1.20, s | 167.4, C | |
| 20 | 55.3, CH3 | 3.80, s | ||
| 13-OH | 9.40, s | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Jiang, M.; Chen, B.; Salaenoi, J.; Niaz, S.-I.; He, J.; Liu, L. Penicamide A, A Unique N,N′-Ketal Quinazolinone Alkaloid from Ascidian-Derived Fungus Penicillium sp. 4829. Mar. Drugs 2019, 17, 522. https://doi.org/10.3390/md17090522
Chen S, Jiang M, Chen B, Salaenoi J, Niaz S-I, He J, Liu L. Penicamide A, A Unique N,N′-Ketal Quinazolinone Alkaloid from Ascidian-Derived Fungus Penicillium sp. 4829. Marine Drugs. 2019; 17(9):522. https://doi.org/10.3390/md17090522
Chicago/Turabian StyleChen, Senhua, Minghua Jiang, Bin Chen, Jintana Salaenoi, Shah-Iram Niaz, Jianguo He, and Lan Liu. 2019. "Penicamide A, A Unique N,N′-Ketal Quinazolinone Alkaloid from Ascidian-Derived Fungus Penicillium sp. 4829" Marine Drugs 17, no. 9: 522. https://doi.org/10.3390/md17090522
APA StyleChen, S., Jiang, M., Chen, B., Salaenoi, J., Niaz, S. -I., He, J., & Liu, L. (2019). Penicamide A, A Unique N,N′-Ketal Quinazolinone Alkaloid from Ascidian-Derived Fungus Penicillium sp. 4829. Marine Drugs, 17(9), 522. https://doi.org/10.3390/md17090522