Antibacterial Polyketides from Antarctica Sponge-Derived Fungus Penicillium sp. HDN151272
Abstract
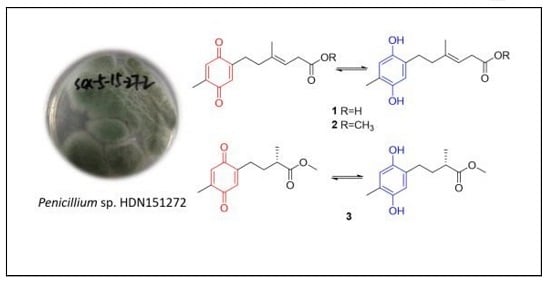
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation
3.4. Extraction and Purification
3.5. Assay of Antimicrobial Activity
3.6. Computation Section
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tian, Y.; Li, Y.L.; Zhao, F.C. Secondary Metabolites from Polar Organisms. Mar. Drugs 2017, 15, 28. [Google Scholar] [CrossRef] [Green Version]
- Bratchkova, A.; Ivanova, V. Bioactive Metabolites Produced by Microorganisms Collected in Antarctica and the Arctic. Biotechnol. Biotech. Equip. 2014, 25, 1–7. [Google Scholar]
- Martinez-Rosales, C.; Fullana, N.; Musto, H.; Castro-Sowinski, S. Antarctic DNA moving forward: Genomic plasticity and biotechnological potential. FEMS Microbiol. Lett. 2012, 331, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wei, X.; Qin, X.; Tian, X.; Liao, L.; Li, K.; Zhou, X.; Yang, X.; Wang, F.; Zhang, T.; et al. Antiviral Merosesquiterpenoids Produced by the Antarctic Fungus Aspergillus ochraceopetaliformis SCSIO 05702. J. Nat. Prod. 2016, 79, 59–65. [Google Scholar]
- Wang, J.; He, W.; Kong, F.; Tian, X.; Wang, P.; Zhou, X.; Liu, Y. Ochracenes A-I, Humulane-Derived Sesquiterpenoids from the Antarctic Fungus Aspergillus ochraceopetaliformis. J. Nat. Prod. 2017, 80, 1725–1733. [Google Scholar] [PubMed]
- Zhou, H.; Li, L.; Wu, C.; Kurtan, T.; Mandi, A.; Liu, Y.; Gu, Q.; Zhu, T.; Guo, P.; Li, D. Penipyridones A-F, Pyridone Alkaloids from Penicillium funiculosum. J. Nat. Prod. 2016, 79, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Zhang, Z.Z.; Feng, Y.Y.; Gu, Q.Q.; Li, D.H.; Zhu, T.J. Secondary metabolites from Antarctic marine-derived fungus Penicillium crustosum HDN153086. Nat. Prod. Res. 2019, 33, 414–419. [Google Scholar] [PubMed]
- Zhou, H.; Li, L.; Wang, W.; Che, Q.; Li, D.; Gu, Q.; Zhu, T. Chrodrimanins I and J from the Antarctic Moss-Derived Fungus Penicillium funiculosum GWT2-24. J. Nat. Prod. 2015, 78, 1442–1445. [Google Scholar]
- Li, L.; Li, D.; Luan, Y.; Gu, Q.; Zhu, T. Cytotoxic metabolites from the antarctic psychrophilic fungus Oidiodendron truncatum. J. Nat. Prod. 2012, 75, 920–927. [Google Scholar] [CrossRef]
- Zhang, T.; Zhu, M.L.; Sun, G.Y.; Li, N.; Gu, Q.Q.; Li, D.H.; Che, Q.; Zhu, T.J. Exopisiod B and farylhydrazone C, two new alkaloids from the Antarctic-derived fungus Penicillium sp. HDN14-431. J. Asian Nat. Prod. Res. 2016, 18, 959–965. [Google Scholar] [CrossRef]
- Capon, R.J.; Ghisalberti, E.L.; Jefferies, P.R. Isoprenoid dihydroquinones from a brown alga, Cystophora sp. Phytochemistry 1981, 20, 2598–2600. [Google Scholar] [CrossRef]
- Reddy, P.; Urban, S. Meroditerpenoids from the southern Australian marine brown alga Sargassum fallax. Phytochemistry 2009, 70, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Li, Y.; Fu, S.; Liu, S.; Wei, J.; Che, Y. Ambuic Acid and Torreyanic Acid Derivatives from the Endolichenic Fungus Pestalotiopsis sp. J. Nat. Prod. 2009, 72, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Spartan’14; Wavefunction Inc.: Irvine, CA, USA, 2013; Available online: https://downloadly.ir/software/engineering-specialized/wavefuncion-spartan/ (accessed on 22 January 2020).
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision, A.1; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Bruhn, T.; Hemberger, Y.; Schauml’ffel, A.; Bringmann, G. SpecDis, Version 1.60; University of Wuerzburg: Wuerzburg, Germany, 2011. [Google Scholar]
- Asche, C. Antitumour quinones. Mini-Rev. Med. Chem. 2005, 5, 449–467. [Google Scholar] [CrossRef]
- Cimino, G.; De Stefano, S.T.; Minale, L. Polyprenyl derivatives from the sponge Ircinia spinosula: 2-Polyprenylbenzoquinones, 2-polyprenylbenzoquinols, prenylated furans and a C-31 difuranoterpene. Tetrahedron 1972, 28, 1315–1324. [Google Scholar] [CrossRef]
- Minale, L.; Riccio, R.; Sodano, G. Avarol a novel sesquiterpenoid hydroquinone with a rearranged drimane skeleton from the sponge Disidea Avara. Tetrahedron Lett. 1974, 15, 3401–3404. [Google Scholar] [CrossRef]
- Laird, D.W.; van Altena, I.A. Tetraprenyltoluquinols from the brown alga Cystophora fibrosa. Phytochemistry 2006, 67, 944–955. [Google Scholar] [CrossRef]
- Mori, J.; Iwashima, M.; Wakasugi, H.; Saito, H.; Matsunaga, T.; Ogasawara, M.; Takahashi, S.; Suzuki, H.; Hayashi, T. New plastoquinones isolated from the brown alga, Sargassum micracanthum. Chem. Pharm. Bul. 2005, 53, 1159–1163. [Google Scholar] [CrossRef] [Green Version]
- Mori, J.; Hayashi, T.; Iwashima, M.; Matsunaga, T.; Saito, H. Effects of plastoquinones from the brown alga Sargassum micracanthum and a new chromene derivative converted from the plastoquinones on acute gastric lesions in rats. Biol. Pharm. Bull. 2006, 29, 1197–1201. [Google Scholar] [CrossRef] [Green Version]
- Aknin, M.; Dayan, T.L.-A.; Rudi, A.; Kashman, Y.; Gaydou, E.M. Hydroquinone antioxidants from the Indian Ocean tunicate Aplidium savignyi. J. Agric. Food Chem. 1999, 47, 4175–4177. [Google Scholar] [CrossRef]
- Appleton, D.R.; Chuen, C.S.; Berridge, M.V.; Webb, V.L.; Copp, B.R. Rossinones A and B, biologically active meroterpenoids from the Antarctic ascidian, Aplidium species. J. Org. Chem. 2009, 74, 9195–9198. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.T.; Pearce, A.N.; Januario, A.H.; Page, M.J.; Kaiser, M.; McLaughlin, R.J.; Harper, J.L.; Webb, V.L.; Barker, D.; Copp, B.R. Anti-inflammatory and antimalarial meroterpenoids from the New Zealand ascidian Aplidium scabellum. J. Org. Chem. 2011, 76, 9151–9156. [Google Scholar] [CrossRef] [PubMed]
- Son, B.W.; Kim, J.C.; Choi, H.D.; Kang, J.S. A radical scavenging Farnesylhydroquinone from a marine-derived fungus Penicillium sp. Arch. Pharm. Res. 2002, 25, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Choi, H.D.; Kang, J.S.; Lee, C.-O.; Son, B.W. New polyoxygenated farnesylcyclohexenones, deacetoxyyanuthone A and its hydro derivative from the marine-derived fungus Penicillium sp. J. Nat. Prod. 2003, 66, 1499–1500. [Google Scholar] [CrossRef] [PubMed]
- Fisch, K.M.; Böhm, V.; Wright, A.D.; König, G.M. Antioxidative Meroterpenoids from the Brown Alga Cystoseira c rinita. J. Nat. Prod. 2003, 66, 968–975. [Google Scholar] [CrossRef] [PubMed]





| No. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| 1H | 13C (Type) | 1H | 13C (Type) | 1H | 13C (Type) | |
| 1 | 147.5, C | 147.5, C | 147.5, C | |||
| 2 | 126.0, C | 125.9, C | 125.4, C | |||
| 3 | 6.44, s | 116.2, CH | 6.44, s | 117.4, CH | 6.41, s | 116.2, CH |
| 4 | 147.9, C | 147.9, C | 147.9, C | |||
| 5 | 121.7, C | 121.8, C | 121.9, C | |||
| 6 | 6.44, s | 117.5, CH | 6.43, s | 116.3, CH | 6.44, s | 117.5, CH |
| 7 | 2.48, m | 28.7, CH2 | 2.46, m | 28.6, CH2 | 2.36, ov. | 27.3, CH2 |
| 8 | 2.12, m | 39.9, CH2 | 2.13, t (8.5) | 39.8, CH2 | 1.53, m | 33.9, CH2 |
| 1.75, m | ||||||
| 9 | 138.2, C | 138.9, C | 2.36, ov. | 38.6, CH | ||
| 10 | 5.26, t (7.2) | 116.8, CH | 5.24, m | 116.1, CH | 176.7, C | |
| 11 | 2.93, d (7.1) | 33.7, CH2 | 3.03, d (7.1) | 33.3, CH2 | 3.58, s | 51.7, CH3 |
| 12 | 173.5, C | 172.4, C | 1.97, s | 16.2, CH3 | ||
| 13 | 3.57, s | 51.8, CH3 | 1.08, s | 17.2, CH3 | ||
| 14 | 1.60, s | 16.7, CH3 | 1.61, s | 16.7, CH3 | ||
| 15 | 1.97, s ov. | 16.2,CH3 | 1.97, s | 16.2, CH3 | ||
| 1-OH | 8.34, s | 8.35, s | 8.35, s | |||
| 4-OH | 8.30, s | 8.31, s | 8.33, s | |||
| No. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| a | b | a | b | a | b | |
| 3 | 6.53, ov. | 6.53, ov. | 6.55, ov. | 6.55, ov. | 6.55, s | 6.52, s |
| 6 | 6.53, ov. | 6.53, ov. | 6.55, ov. | 6.55, ov. | 6.59, s | 6.62, s |
| 7 | 2.51, t (7.5) | 2.62, m | 2.53, t (7.4) | 2.65, t (7.5) | 2.42, m | 2.52, ov. |
| 8 | 2.20, t (7.5) | 2.26, t (7.8) | 2.23, t (7.4) | 2.28, t (7.5) | 1.88, ov. | 1.89, ov. |
| 1.62, m | 1.71, m | |||||
| 9 | 2.51, ov. | 2.51, ov. | ||||
| 10 | 5.32, ov. | 5.28, ov. | 5.32, t (6.4) | 5.28,t (6.9) | ||
| 11 | 3.02, ov. | 3.02, ov. | 3.04, ov. | 3.04, ov. | 3.68, s | 3.72, s |
| 12 | 2.04, s | 2.17, s | ||||
| 13 | 3.67, s | 3.68, s | 1.19, ov. | 1.20, ov. | ||
| 14 | 2.00, s | 2.12, s | 2.03, s | 2.16, s | ||
| 15 | 1.64, s | 1.65, s | 1.66, s | 1.68, s | ||
| No. | 1 (Type) | 2 (Type) | 3 (Type) | |||
|---|---|---|---|---|---|---|
| a | b | a | b | a | b | |
| 1 | 147.6, C | 185.9, C | 146.9, C | 187.6, C | 147.7, C | 187.5, C |
| 2 | 127.0, C | 148.8, C | 126.7, C | 148.7, C | 125.6, C | 148.6, C |
| 3 | 117.6, CH | 132.9, CH | 116.7, CH | 132.8, CH | 116.3, CH | 132.7, CH |
| 4 | 147.7, C | 187.7, C | 147.6, C | 188.1, C | 147.3, C | 188.1, C |
| 5 | 122.1, C | 145.6, C | 122.2, C | 145.6, C | 122.7, C | 145.7, C |
| 6 | 118.0, CH | 133.6, CH | 118.0, CH | 133.5, CH | 118.3, CH | 133.6, CH |
| 7 | 29.8, CH2 | 27.2, CH2 | 27.9, CH2 | 27.1, CH2 | 27.6, CH2 | 26.5, CH2 |
| 8 | 39.8, CH2 | 37.7, CH2 | 39.7, CH2 | 37.6, CH2 | 34.0, CH2 | 31.5, CH2 |
| 9 | 138.2, C | 137.3, C | 139.2, C | 137.3, C | 38.7, CH | 39.0, CH |
| 10 | 116.9, CH | 118.0, CH | 116.3, CH | 117.2, CH | 177.9, C | 176.5, C |
| 11 | 33.6, CH2 | 33.4, CH2 | 33.5, CH2 | 33.5, CH2 | 52.0, CH3 | 51.7, CH3 |
| 12 | 173.7, C | 171.9, C | 173.2, C | 172.5, C | 15.5, CH3 | 15.5, CH3 |
| 13 | 51.9, CH3 | 51.8, CH3 | 17.3, CH3 | 17.1, CH3 | ||
| 14 | 15.5, CH3 | 15.7, CH3 | 15.4, CH3 | 15.5, CH3 | ||
| 15 | 16.7, CH3 | 16.3, CH3 | 16.5, CH3 | 16.2, CH3 | ||
| Compd. | V. Parahemolyticus | E. coli | Prot-eus sp. | B. subtilis | MRCNS | B. cereus | P. aeruginosa | M. Phlei | M. albican |
|---|---|---|---|---|---|---|---|---|---|
| 1 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 2 | >50 | >50 | >50 | >50 | 6.25 | 12.50 | 1.56 | 3.13 | >50 |
| 3 | 12.50 | >50 | >50 | 12.50 | 6.25 | 25.00 | 6.25 | 6.25 | >50 |
| Ciprofloxacin | 0.52 | 2.07 | 0.52 | 0.98 | 0.98 | 0.98 | 0.98 | 3.91 | 3.91 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, M.; Sun, C.; Sun, Z.; Zhang, G.; Che, Q.; Gu, Q.; Zhu, T.; Li, D. Antibacterial Polyketides from Antarctica Sponge-Derived Fungus Penicillium sp. HDN151272. Mar. Drugs 2020, 18, 71. https://doi.org/10.3390/md18020071
Shah M, Sun C, Sun Z, Zhang G, Che Q, Gu Q, Zhu T, Li D. Antibacterial Polyketides from Antarctica Sponge-Derived Fungus Penicillium sp. HDN151272. Marine Drugs. 2020; 18(2):71. https://doi.org/10.3390/md18020071
Chicago/Turabian StyleShah, Mudassir, Chunxiao Sun, Zichao Sun, Guojian Zhang, Qian Che, Qianqun Gu, Tianjiao Zhu, and Dehai Li. 2020. "Antibacterial Polyketides from Antarctica Sponge-Derived Fungus Penicillium sp. HDN151272" Marine Drugs 18, no. 2: 71. https://doi.org/10.3390/md18020071
APA StyleShah, M., Sun, C., Sun, Z., Zhang, G., Che, Q., Gu, Q., Zhu, T., & Li, D. (2020). Antibacterial Polyketides from Antarctica Sponge-Derived Fungus Penicillium sp. HDN151272. Marine Drugs, 18(2), 71. https://doi.org/10.3390/md18020071