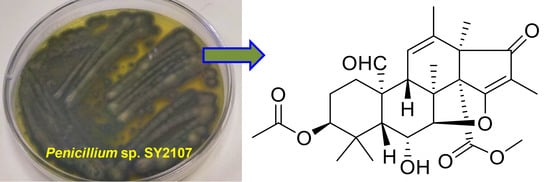
Bioactive Metabolites from the Mariana Trench Sediment-Derived Fungus Penicillium sp. SY2107
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Isolation and Identification of Strain SY2107
3.3. Scale Up Culture of Strain SY2107
3.4. Isolation of Compounds 1–16
3.5. Antimicrobial Active Assay
3.6. Antiproliferative Active Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [Green Version]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, R.B.; Evdokimov, N.M.; Lefranc, F.; Valentão, P.; Kornienko, A.; Pereira, D.M.; Andrade, P.B.; Gomes, N.G.M. Marine-derived anticancer agents: Clinical benefits, innovative mechanisms, and new targets. Mar. Drugs 2019, 17, 329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinde, P.; Banerjee, P.; Mandhare, A. Marine natural products as source of new drugs: A patent review (2015–2018). Expert Opin. Ther. Pat. 2019, 29, 283–309. [Google Scholar] [CrossRef] [PubMed]
- Matulja, D.; Wittine, K.; Malatesti, N.; Laclef, S.; Turks, M.; Kolympadi, M.; Ambrožić, G.; Markovic, D. Marine natural products with high anticancer activities. Curr. Med. Chem. 2020, 27, 1243–1307. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D. Deep-sea natural products. Nat. Prod. Rep. 2008, 25, 1131–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Pilkington, L.I. A Chemometric analysis of deep-sea natural products. Molecules 2019, 24, 3942. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Mudassir, S.; Zhang, Z.; Feng, Y.; Chang, Y.; Che, Q.; Gu, Q.; Zhu, T.; Zhang, G.; Li, D. Secondary metabolites from deep-sea derived microorganisms. Curr. Med. Chem. 2019. [Google Scholar] [CrossRef]
- Prieto-Davó, A.; Villarreal-Gómez, L.J.; Forschner-Dancause, S.; Bull, A.T.; Stach, J.E.; Smith, D.C.; Rowley, D.C.; Jensen, P.R. Targeted search for actinomycetes from nearshore and deep-sea marine sediments. FEMS Microbiol. Ecol. 2013, 84, 510–518. [Google Scholar] [CrossRef]
- Russo, P.; Del Bufalo, A.; Fini, M. Deep sea as a source of novel-anticancer drugs: Update on discovery and preclinical/clinical evaluation in a systems medicine perspective. EXCLI J. 2015, 14, 228–236. [Google Scholar] [PubMed]
- Jensen, P.R.; Moore, B.S.; Fenical, W. The marine actinomycete genus Salinispora: A model organism for secondary metabolite discovery. Nat. Prod. Rep. 2015, 32, 738–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di, K.; Lloyd, G.K.; Abraham, V.; MacLaren, A.; Burrows, F.J.; Desjardins, A.; Trikha, M.; Bota, D.A. Marizomib activity as a single agent in malignant gliomas: Ability to cross the blood-brain barrier. Neuro Oncol. 2016, 18, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Badros, A.; Singh, Z.; Dhakal, B.; Kwok, Y.; MacLaren, A.; Richardson, P.; Trikha, M.; Hari, P. Marizomib for central nervous system-multiple myeloma. Br. J. Haematol. 2017, 177, 221–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peoples, L.M.; Grammatopoulou, E.; Pombrol, M.; Xu, X.; Osuntokun, O.; Blanton, J.; Allen, E.E.; Nunnally, C.C.; Drazen, J.C.; Mayor, D.J.; et al. Microbial community diversity within sediments from two geographically separated hadal trenches. Front. Microbiol. 2019, 10, 347. [Google Scholar] [CrossRef]
- Liu, J.; Zheng, Y.; Lin, H.; Wang, X.; Li, M.; Liu, Y.; Yu, M.; Zhao, M.; Pedentchouk, N.; Lea-Smith, D.J.; et al. Proliferation of hydrocarbon-degrading microbes at the bottom of the Mariana Trench. Microbiome 2019, 7, 47. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Chai, W.Y.; Wang, W.L.; Song, T.F.; Lian, X.Y.; Zhang, Z.Z. Cytotoxic bagremycins from mangrove-derived Streptomyces sp. Q22. J. Nat. Prod. 2017, 80, 1450–1456. [Google Scholar] [CrossRef]
- Chen, M.X.; Chai, W.Y.; Song, T.F.; Ma, M.Z.; Lian, X.Y.; Zhang, Z.Z. Anti-glioma natural products downregulating tumor glycolytic enzymes from marine actinomycete Streptomyces sp. ZZ406. Sci. Rep. 2018, 8, 72. [Google Scholar] [CrossRef] [Green Version]
- Song, T.F.; Chen, M.X.; Ge, Z.W.; Chai, W.Y.; Li, X.C.; Zhang, Z.Z.; Lian, X.Y. Bioactive penicipyrrodiether A, an adduct of GKK1032 analogue and phenol A derivative, from a marine-sourced fungus Penicillium sp. ZZ380. J. Org. Chem. 2018, 83, 13395–13401. [Google Scholar] [CrossRef]
- Song, T.F.; Tang, M.M.; Ge, H.J.; Chen, M.X.; Lian, X.Y.; Zhang, Z.Z. Novel bioactive penicipyrroether A and pyrrospirone J from the marine-derived Penicillium sp. ZZ380. Mar. Drugs 2019, 17, 292. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Yi, W.W.; Ge, H.Z.; Zhang, Z.Z.; Wu, B. Bioactive streptoglutarimides A–J from the marine-derived Streptomyces sp. ZZ741. J. Nat. Prod. 2019, 82, 2800–2808. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.W.; Xie, C.L.; Xia, J.M.; Luo, Q.; Luo, Z.H.; Shao, Z.Z. Heteroterpene Compound Derived from Penicillium and Its Preparation Method for Anti-Tumor Medicament. CN Patent 109942658 A 20190628, 2019. [Google Scholar]
- Xie, C.L.; Xia, J.M.; Lin, T.; Lin, Y.J.; Lin, Y.K.; Xia, M.L.; Chen, H.F.; Luo, Z.H.; Shao, Z.Z.; Yang, X.W. Andrastone A from the deep-sea-derived fungus Penicillium allii-sativi acts as an inducer of Caspase and RXRα-dependent apoptosis. Front. Chem. 2019, 7, 692. [Google Scholar] [CrossRef] [PubMed]
- Umehara, K.; Yoshida, K.; Okamoto, M.; Iwami, M.; Tanaka, H.; Kohsaka, M.; Imanaka, H. Studies on WF-5239, a new potent platelet aggregation inhibitor. J. Antibiot. 1984, 37, 469–474. [Google Scholar] [CrossRef] [Green Version]
- Qu, P.; Liu, P.P.; Fu, P.; Wang, Y.; Zhu, W.M. Secondary metabolites of halotolerant fungus Penicillium chrysogenum HK14-01 from the Yellow River Delta area. Acta Microbiol. Sin. 2012, 52, 1103–1112. [Google Scholar]
- Wang, C.; Gao, Y.K.; Lei, F.H.; Tan, X.C.; Shen, L.Q.; Gao, C.H.; Yi, X.X.; Li, X.Y. A new glycosyl ester isolated from marine-derived Penicillium sp. Chin. Trad. Herbal Drugs 2019, 50, 2518–2523. [Google Scholar]
- Takahashi, C.; Matsushita, T.; Doi, M.; Minoura, K.; Shingu, T.; Kumeda, Y.; Numata, A. Fumiquinazolines A–G, novel metabolites of a fungus separated from a Pseudolabrus marine fish. J. Chem. Soc. Perkin Trans. 1995, 18, 2345–2353. [Google Scholar] [CrossRef]
- Wang, F.Z.; Fang, Y.C.; Zhu, T.J.; Zhang, M.; Lin, A.Q.; Gu, Q.Q.; Zhu, W.M. Seven new prenylated indole diketopiperazine alkaloids from holothurian-derived fungus Aspergillus Fumigatus. Tetrahedron 2008, 64, 7986–7991. [Google Scholar] [CrossRef]
- Zhang, L.M.; Li, Z.L.; Bai, J.; Wu, X.; Wang, Y.; Hua, H.M. Metabolites of Aspergillus sp. HT-2. Chin. Pharm. J. 2011, 46, 1154–1158. [Google Scholar]
- Peter, B.; Christoph, T. Isolation and structure of pseurotin A, a microbial metabolite of Pseudeurotium ovalis Stolk with an unusual heterospirocyclic system. Helvetica Chim. Acta 1981, 64, 304–315. [Google Scholar]
- Lin, X.P.; Wu, Q.Y.; Yu, Y.Y.; Liang, Z.; Liu, Y.H.; Zhou, L.L.; Tang, L.; Zhou, X.F. Penicilliumin B, a novel sesquiterpene methylcyclopentenedione from a deep sea-derived Penicillium strain with renoprotective activities. Sci. Rep. 2017, 7, 10757. [Google Scholar] [CrossRef] [Green Version]
- Ji, Y.; Xin, Z.; He, H.; Gao, S. Total synthesis of viridin and viridiol. J. Am. Chem. Soc. 2019, 141, 16208–16212. [Google Scholar] [CrossRef]
- Jongrungruangchok, S.; Kittakoop, P.; Yongsmith, B.; Bavovada, R.; Tanasupawat, S.; Lartpornmatulee, N.; Thebtaranonth, Y. Azaphilone pigments from a yellow mutant of the fungus Monascus kaoliang. Phytochemistry 2004, 65, 2569–2575. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wei, M.; Chen, G.; Lin, Y. Two new dihydroisocoumarins from the endophytic fungus Aspergillus sp. collected from the South China Sea. Chem. Nat. Comp. 2012, 48, 371–373. [Google Scholar] [CrossRef]
- Zuo, M.X.; Zhou, Y.L.; Xu, Y.C.; Liu, W.; Zhu, W.M.; Wang, L.P. Secondary metabolites of Aspergillus fumigatus GZWMJZ-152 cultivated on Camellia seed cake medium. Mycosystema 2019, 38, 264–271. [Google Scholar]
- Liu, R.; Zhu, W.M.; Zhang, Y.P.; Zhu, T.J.; Liu, H.B.; Fang, Y.C.; Gu, Q.Q. A new diphenyl ether from marine-derived fungus Aspergillus sp. B-F-2. J. Antibiot. 2006, 59, 362–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, X.C.; Zhuang, Y.B.; Wang, Y.; Liu, P.P.; Xu, Z.H.; Zhu, W.M. Secondary metabolites from Penicillium sp. gxwz406 symbiotic with the gorgonian Echinogorgia flora. Chin. J. Mar. Drugs 2010, 29, 11–15. [Google Scholar]
- Fu, H.C.; Zhao, W.Y.; Zhong, H.M.; Hong, K.; Gu, Q.Q.; Zhu, W.M. Bioactive alkaloids from sponge-derived actinomyces sh6004. Chin. J. Mar. Drugs 2008, 27, 14–20. [Google Scholar]
- Ye, X.W.; Anjum, K.; Song, T.F.; Wang, W.L.; Yu, S.R.; Huang, H.C.; Lian, X.Y.; Zhang, Z.Z. A new curvularin glycoside and its cytotoxic and antibacterial analogues from marine actinomycete Pseudonocardia sp. HS7. Nat. Prod. Res. 2016, 30, 1156–1161. [Google Scholar] [CrossRef]
- Xin, W.X.; Ye, X.W.; Yu, S.R.; Lian, X.Y.; Zhang, Z.Z. New capoamycin-type antibiotics and polyene acids from marine Streptomyces fradiae PTZ0025. Mar. Drugs 2012, 10, 2388–2402. [Google Scholar] [CrossRef] [Green Version]



| No. | 13C, Type | 1H (J in Hz) | No. | 13C, Type | 1H (J in Hz) |
|---|---|---|---|---|---|
| 1 | 28.5, CH2 | α: 2.31 dt (13.5, 3.2); β: 0.79, td (13.5, 4.5) | 15 | 111.6, C | – |
| 2 | 22.4, CH2 | 1.47, m | 16 | 185.3, C | – |
| 3 | 77.4, CH | 4.42, t (3.1) | 17 | 68.5, C | – |
| 4 | 36.8, C | – | 18 | 25.3, CH3 | 0.97, s |
| 5 | 45.8, CH | 1.79, d (2.8) | 19 | 23.0, CH3 | 1.09, s |
| 6 | 66.0, CH | 4.52, t (2.8) | 20 | 15.5, CH3 | 1.12, s |
| 7 | 91.9, CH | 4.67, d (2.8) | 21 | 207.7, CH | 10.55, s |
| 8 | 45.6, C | – | 22 | 20.2, CH3 | 1.75, s |
| 9 | 50.1, CH | 2.05, br s | 23 | 20.5, CH3 | 1.13, s |
| 10 | 51.5, C | – | 24 | 5.4, CH3 | 1.57, s |
| 11 | 120.5, CH | 5.60, br s | 25 | 168.3, C | – |
| 12 | 133.9, C | – | 26 | 169.6, C | – |
| 13 | 52.9, C | – | 27 | 20.6, CH3 | 1.93, s |
| 14 | 201.1, C | – | 28 | 52.4, CH3 | 3.69, s |
| Compounds | MRSA | E. coli | Candida albicans | |||
|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | |
| 1 | 8 | 15 | 8 | 13 | 13 | 17 |
| 2 | 9 | 15 | 12 | 19 | 6 | 10 |
| 3 | 13 | 18 | 10 | 20 | 9 | 16 |
| 4 | 14 | 20 | 13 | 18 | 14 | 20 |
| 5 | 15 | 22 | 9 | 16 | 12 | 18 |
| 6 | 11 | 20 | 9 | 14 | 13 | 20 |
| 7 | 15 | 20 | 16 | 23 | 15 | 21 |
| 8 | 17 | 23 | 16 | 22 | 10 | 18 |
| 9 | 12 | 18 | 13 | 21 | 15 | 24 |
| 10 | 20 | 26 | 22 | 28 | 14 | 26 |
| 11 | 9 | 15 | 11 | 19 | 17 | 28 |
| 12 | 15 | 26 | 14 | 23 | 18 | 28 |
| 13 | 28 | 36 | 22 | 30 | >50 | >50 |
| 14 | 33 | 39 | 38 | 42 | >50 | >50 |
| 15 | 32 | 41 | 34 | 45 | >50 | >50 |
| 16 | 27 | 33 | 26 | 32 | >50 | >50 |
| Gentamicin | 3 | 7 | 0.5 | 3 | NT | NT |
| Vancomycin | 0.5 | 3 | NT | NT | NT | NT |
| Amphotericin B | NT | NT | NT | NT | 3 | 6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaleem, S.; Qin, L.; Yi, W.; Lian, X.-Y.; Zhang, Z. Bioactive Metabolites from the Mariana Trench Sediment-Derived Fungus Penicillium sp. SY2107. Mar. Drugs 2020, 18, 258. https://doi.org/10.3390/md18050258
Kaleem S, Qin L, Yi W, Lian X-Y, Zhang Z. Bioactive Metabolites from the Mariana Trench Sediment-Derived Fungus Penicillium sp. SY2107. Marine Drugs. 2020; 18(5):258. https://doi.org/10.3390/md18050258
Chicago/Turabian StyleKaleem, Sidra, Le Qin, Wenwen Yi, Xiao-Yuan Lian, and Zhizhen Zhang. 2020. "Bioactive Metabolites from the Mariana Trench Sediment-Derived Fungus Penicillium sp. SY2107" Marine Drugs 18, no. 5: 258. https://doi.org/10.3390/md18050258
APA StyleKaleem, S., Qin, L., Yi, W., Lian, X. -Y., & Zhang, Z. (2020). Bioactive Metabolites from the Mariana Trench Sediment-Derived Fungus Penicillium sp. SY2107. Marine Drugs, 18(5), 258. https://doi.org/10.3390/md18050258