Bactericidal Activity of Usnic Acid-Chitosan Nanoparticles against Persister Cells of Biofilm-Forming Pathogenic Bacteria
Abstract
:1. Introduction
2. Result and Discussion
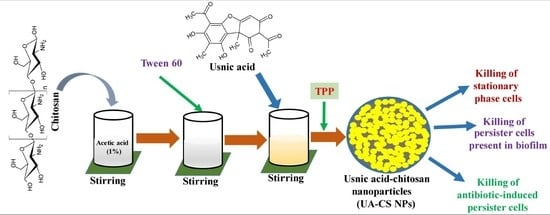
2.1. Preparation and Characterization of UA-CS NPs
2.2. Minimum Inhibitory Concentration (MIC) Values of UA-CS NPs against Gram-Positive and Gram-Negative Bacteria
2.3. Eradication Effects of UA-CS NPs Towards Different Types of Persister Cells
2.3.1. Anti-Persister Activity of UA-CS NPs Towards Stationary-Phase Cultures
2.3.2. Anti-Persister Activity of UA-CS NPs Towards the Biofilm-Associated Persister Cells
2.3.3. Anti-Persister Activity of UA-CS NPs towards Antibiotic-Induced Cells
2.4. Effect of UA-CS NPs on Cell Morphology
3. Conclusions
4. Materials and Methods
4.1. Bacterial Strains, Growth Conditions, and Reagents
4.2. Preparation of Usnic Acid-Chitosan Nanoparticles (UA-CS NPs)
4.3. Characterization of UA-CS NPs
4.4. Determination of Encapsulation Efficiency and Loading Capacity
4.5. In Vitro Release of Usnic Acid from UA-CS NPs
4.6. Minimum Inhibitory Concentration (MIC) Determination of UA-CS NPs
4.7. Measurement of Persister Cells Formation
4.8. Effect of UA-CS NPs on Stationary Phase Cells
4.9. Eradication of Biofilm-Associated Persister Cells by UA-CS NPs
4.10. Eradication of Antibiotic-Induced Persister Cells by UA-CS NPs
4.11. Analysis of Cell Morphology of Bacterial Cells Treated by UA-CS NPs
4.12. Statistical Analysis
4.13. Abbreviations
Author Contributions
Funding
Conflicts of Interest
References
- Maisonneuve, E.; Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 2014, 157, 539–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 2008, 322, 107–131. [Google Scholar] [PubMed]
- Defraine, V.; Fauvart, M.; Michiels, J. Fighting bacterial persistence: Current and emerging anti-persister strategies and therapeutics. Drug Resist. Updates 2018, 38, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells and the riddle of biofilm survival. Biochemistry 2005, 70, 267–274. [Google Scholar] [CrossRef]
- Leszczynska, D.; Matuszewska, E.; Kuczynska-Wisnik, D.; Furmanek-Blaszk, B.; Laskowska, E. The formation of persister cells in stationary-phase cultures of Escherichia coli is associated with the aggregation of endogenous proteins. PLoS ONE 2013, 8, e54737. [Google Scholar] [CrossRef] [Green Version]
- Shan, Y.; Brown Gandt, A.; Rowe, S.E.; Deisinger, J.P.; Conlon, B.P.; Lewis, K. ATP-Dependent persister formation in Escherichia coli. mBio 2017, 8, e02267-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spoering, A.L.; Lewis, K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 2001, 183, 6746–6751. [Google Scholar] [CrossRef] [Green Version]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.-J.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral. Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Khan, F.; Pham, D.T.N.; Oloketuyi, S.F.; Manivasagan, P.; Oh, J.; Kim, Y.M. Chitosan and their derivatives: Antibiofilm drugs against pathogenic bacteria. Colloids Surf. B Biointerfaces 2020, 185, 110627. [Google Scholar] [CrossRef]
- Manivasagan, P.; Oh, J. Marine polysaccharide-based nanomaterials as a novel source of nanobiotechnological applications. Int. J. Biol. Macromol. 2016, 82, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhang, S.; Shi, W.; Zubcevik, N.; Miklossy, J.; Zhang, Y. Selective essential oils from spice or culinary herbs have high activity against stationary phase and biofilm Borrelia burgdorferi. Front. Med. 2017, 4, 169. [Google Scholar] [CrossRef] [Green Version]
- Francolini, I.; Norris, P.; Piozzi, A.; Donelli, G.; Stoodley, P. Usnic acid, a natural antimicrobial agent able to inhibit bacterial biofilm formation on polymer surfaces. Antimicrob. Agents Ch 2004, 48, 4360–4365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciąg-Dorszyńska, M.; Węgrzyn, G.; Guzow-Krzemińska, B. Antibacterial activity of lichen secondary metabolite usnic acid is primarily caused by inhibition of RNA and DNA synthesis. FEMS Microbiol. Lett. 2014, 353, 57–62. [Google Scholar] [CrossRef]
- Tay, T.; Türk, A.O.; Yilmaz, M.; Türk, H.; Kivanç, M. Evaluation of the antimicrobial activity of the acetone extract of the lichen Ramalina farinacea and its (+)-usnic acid, norstictic acid, and protocetraric acid constituents. Z. Nat. C J. Biosci. 2004, 59, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Dai, F.; Zhai, D.; Dong, Y.; Zhang, J.; Lu, B.; Luo, J.; Liu, M.; Yi, Z. Usnic acid inhibits breast tumor angiogenesis and growth by suppressing VEGFR2-mediated AKT and ERK1/2 signaling pathways. Angiogenesis 2012, 15, 421–432. [Google Scholar] [CrossRef]
- Nunes, P.S.; Albuquerque, R.L., Jr.; Cavalcante, D.R.; Dantas, M.D.; Cardoso, J.C.; Bezerra, M.S.; Souza, J.C.; Serafini, M.R.; Quitans, L.J., Jr.; Bonjardim, L.R.; et al. Collagen-based films containing liposome-loaded usnic acid as dressing for dermal burn healing. J. Biomed. Biotechnol. 2011, 2011, 761593. [Google Scholar] [CrossRef]
- Erba, E.; Pocar, D.; Rossi, L.M. New esters of R-(+)-usnic acid. Il Farmaco 1998, 53, 718–720. [Google Scholar] [CrossRef]
- Dasgupta, Q.; Madras, G.; Chatterjee, K. Controlled release of usnic acid from biodegradable polyesters to inhibit biofilm formation. ACS Biomater. Sci. Eng. 2017, 3, 291–303. [Google Scholar] [CrossRef]
- Khan, F.; Khan, M.M.; Kim, Y.M. Recent progress and future perspectives of antibiofilm drugs immobilized on nanomaterials. Curr. Pharm. Biotechnol. 2018, 19, 631–643. [Google Scholar] [CrossRef]
- Keawchaoon, L.; Yoksan, R. Preparation, characterization and in vitro release study of carvacrol-loaded chitosan nanoparticles. Colloids Surf. B Biointerfaces 2011, 84, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Woranuch, S.; Yoksan, R. Eugenol-loaded chitosan nanoparticles: I. Thermal stability improvement of eugenol through encapsulation. Carbohydr. Polym. 2013, 96, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Nedovic, V.; Kalusevic, A.; Manojlovic, V.; Levic, S.; Bugarski, B. An overview of encapsulation technologies for food applications. Proc. Food Sci. 2011, 1, 1806–1815. [Google Scholar] [CrossRef] [Green Version]
- Sowasod, N.; Nakagawa, K.; Tanthapanichakoon, W.; Charinpanitkul, T. Development of encapsulation technique for curcumin loaded O/W emulsion using chitosan based cryotropic gelation. Mater. Sci. Eng. C 2012, 32, 790–798. [Google Scholar] [CrossRef]
- Jung, W.J.; Park, R.D. Bioproduction of chitooligosaccharides: Present and perspectives. Mar. Drugs 2014, 12, 5328–5356. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Yang, W.; Wang, C.; Hu, J.; Fu, S. Chitosan nanoparticles as a novel delivery system for ammonium glycyrrhizinate. Int. J. Pharm. 2005, 295, 235–245. [Google Scholar] [CrossRef]
- Chen, L.; Subirade, M. Chitosan/β-lactoglobulin core–shell nanoparticles as nutraceutical carriers. Biomaterials 2005, 26, 6041–6053. [Google Scholar] [CrossRef]
- Janes, K.A.; Calvo, P.; Alonso, M.J. Polysaccharide colloidal particles as delivery systems for macromolecules. Adv. Drug Deliv. Rev. 2001, 47, 83–97. [Google Scholar] [CrossRef]
- Wilson, B.; Samanta, M.K.; Santhi, K.; Kumar, K.P.; Ramasamy, M.; Suresh, B. Chitosan nanoparticles as a new delivery system for the anti-Alzheimer drug tacrine. Nanomedicine 2010, 6, 144–152. [Google Scholar] [CrossRef]
- Bayat, A.; Dorkoosh, F.A.; Dehpour, A.R.; Moezi, L.; Larijani, B.; Junginger, H.E.; Rafiee-Tehrani, M. Nanoparticles of quaternized chitosan derivatives as a carrier for colon delivery of insulin: Ex vivo and in vivo studies. Int. J. Pharm. 2008, 356, 259–266. [Google Scholar] [CrossRef]
- Araújo, E.S.; Pereira, E.C.; da Costa, M.M.; da Silva, N.H.; de Oliveira, H.P. Bactericidal activity of usnic acid-loaded electrospun fibers. Rec. Pat. Nanotechnol. 2016, 10, 252–257. [Google Scholar] [CrossRef]
- Takani, M.; Yajima, T.; Masuda, H.; Yamauchi, O. Spectroscopic and structural characterization of copper(II) and palladium(II) complexes of a lichen substance usnic acid and its derivatives. Possible forms of environmental metals retained in lichens. J. Inorg. Biochem. 2002, 91, 139–150. [Google Scholar] [CrossRef]
- Bjerke, J.W.; Lerfall, K.; Elvebakk, A. Effects of ultraviolet radiation and PAR on the content of usnic and divaricatic acids in two arctic-alpine lichens. Photochem. Photobiol. Sci. 2002, 1, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Li, P.-W.; Wang, G.; Yang, Z.-M.; Duan, W.; Peng, Z.; Kong, L.-X.; Wang, Q.-H. Development of drug-loaded chitosan–vanillin nanoparticles and its cytotoxicity against HT-29 cells. Drug Deliv. 2016, 23, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Mardyani, S.; Chan, W.C.; Kumacheva, E. Design of biocompatible chitosan microgels for targeted pH-mediated intracellular release of cancer therapeutics. Biomacromolecules 2006, 7, 1568–1572. [Google Scholar] [CrossRef]
- Agnihotri, S.A.; Mallikarjuna, N.N.; Aminabhavi, T.M. Recent advances on chitosan-based micro- and nanoparticles in drug delivery. J. Control. Release 2004, 100, 5–28. [Google Scholar] [CrossRef]
- Ahmed, T.A.; Aljaeid, B.M. Preparation, characterization, and potential application of chitosan, chitosan derivatives, and chitosan metal nanoparticles in pharmaceutical drug delivery. Drug Des. Dev. Ther. 2016, 10, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagano, C.; Ceccarini, M.R.; Calarco, P.; Scuota, S.; Conte, C.; Primavilla, S.; Ricci, M.; Perioli, L. Bioadhesive polymeric films based on usnic acid for burn wound treatment: Antibacterial and cytotoxicity studies. Colloid Surf. B Biointerfaces 2019, 178, 488–499. [Google Scholar] [CrossRef]
- Nallamuthu, I.; Devi, A.; Khanum, F. Chlorogenic acid loaded chitosan nanoparticles with sustained release property, retained antioxidant activity and enhanced bioavailability. Asian J. Pharm. Sci. 2015, 10, 203–211. [Google Scholar] [CrossRef] [Green Version]
- Ding, R.-L.; Xie, F.; Hu, Y.; Fu, S.-Z.; Wu, J.-B.; Fan, J.; He, W.-F.; He, Y.; Yang, L.-L.; Lin, S.; et al. Preparation of endostatin-loaded chitosan nanoparticles and evaluation of the antitumor effect of such nanoparticles on the Lewis lung cancer model. Drug Deliv. 2017, 24, 300–308. [Google Scholar] [CrossRef] [Green Version]
- Ajun, W.; Yan, S.; Li, G.; Huili, L. Preparation of aspirin and probucol in combination loaded chitosan nanoparticles and in vitro release study. Carbohydr. Polym. 2009, 75, 566–574. [Google Scholar] [CrossRef]
- Wisuitiprot, W.; Somsiri, A.; Ingkaninan, K.; Waranuch, N. A novel technique for chitosan microparticle preparation using a water/silicone emulsion: Green tea model. Int. J. Cosmet. Sci. 2011, 33, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Alpsoy, L.; Baykal, A.; Amir, M.; Ülker, Z.; Nawaz, M. SPION@APTES@FA-PEG@Usnic acid bionanodrug for cancer therapy. J. Supercond. Nov. Magn. 2018, 31, 1395–1401. [Google Scholar] [CrossRef]
- Fernandes Queiroz, M.; Melo, K.R.; Sabry, D.A.; Sassaki, G.L.; Rocha, H.A. Does the use of chitosan contribute to oxalate kidney stone formation? Mar. Drugs 2014, 13, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Yu, H.; Zhang, M.; Yang, Y.; Zhang, G. Physicochemical properties and antioxidant activity of chitosan from the blowfly Chrysomya megacephala larvae. Int. J. Biol. Macromol. 2013, 60, 347–354. [Google Scholar] [CrossRef]
- Lira, M.C.B.; Ferraz, M.S.; da Silva, D.G.V.C.; Cortes, M.E.; Teixeira, K.I.; Caetano, N.P.; Sinisterra, R.D.; Ponchel, G.; Santos-Magalhães, N.S. Inclusion complex of usnic acid with β-cyclodextrin: Characterization and nanoencapsulation into liposomes. J. Incl. Phenom. Macro. Chem. 2009, 64, 215–224. [Google Scholar] [CrossRef]
- Fitriani, L.; Afriyanti, I.; Afriyani Farmasi, A.; Ismed, F.; Zaini, E. Solid dispersion of usnic acid–HPMC 2910 prepared by spray drying and freeze drying techniques. Orient J. Chem. 2018, 34, 2083–2088. [Google Scholar] [CrossRef]
- Hosseini, S.F.; Zandi, M.; Rezaei, M.; Farahmandghavi, F. Two-step method for encapsulation of oregano essential oil in chitosan nanoparticles: Preparation, characterization and in vitro release study. Carbohydr. Polym. 2013, 95, 50–56. [Google Scholar] [CrossRef]
- Yoksan, R.; Jirawutthiwongchai, J.; Arpo, K. Encapsulation of ascorbyl palmitate in chitosan nanoparticles by oil-in-water emulsion and ionic gelation processes. Colloid Surf. B Biointerfaces 2010, 76, 292–297. [Google Scholar] [CrossRef]
- Al-Nemrawi, N.K.; Okour, A.R.; Dave, R.H. Surface modification of PLGA nanoparticles using chitosan: Effect of molecular weight, concentration, and degree of deacetylation. Adv. Polym. Technol. 2018, 37, 3066–3075. [Google Scholar] [CrossRef]
- Nithyanand, P.; Beema Shafreen, R.M.; Muthamil, S.; Karutha Pandian, S. Usnic acid, a lichen secondary metabolite inhibits Group A Streptococcus biofilms. Antonie Van Leeuwenhoek 2015, 107, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Riviello, A.; Crocetta, V.; Di Giuseppe, F.; Pomponio, S.; Sulpizio, M.; Di Ilio, C.; Angelucci, S.; Barone, L.; Di Giulio, A.; et al. Evaluation of antibacterial and antibiofilm mechanisms by usnic acid against methicillin-resistant Staphylococcus aureus. Future Microbiol. 2016, 11, 1315–1338. [Google Scholar] [CrossRef]
- Francolini, I.; Taresco, V.; Crisante, F.; Martinelli, A.; D’Ilario, L.; Piozzi, A. Water soluble usnic acid-polyacrylamide complexes with enhanced antimicrobial activity against Staphylococcus epidermidis. Int. J. Mol. Sci. 2013, 14, 7356–7369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingólfsdóttir, K.N.; Chung, G.A.C.; Skúlason, V.G.; Gissurarson, S.R.; Vilhelmsdóttir, M. Antimycobacterial activity of lichen metabolites in vitro. Eur. J. Pharm. Sci. 1998, 6, 141–144. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef] [Green Version]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007, 5, 48–56. [Google Scholar] [CrossRef]
- Michiels, J.E.; Van den Bergh, B.; Verstraeten, N.; Michiels, J. Molecular mechanisms and clinical implications of bacterial persistence. Drug Resist. Updates 2016, 29, 76–89. [Google Scholar] [CrossRef]
- Humphreys, G.J.; McBain, A.J. Chapter Fourteen—An introduction to the biology of biofilm recalcitrance. In Biofilms in Infection Prevention and Control; Percival, S.L., Williams, D.W., Randle, J., Cooper, T., Eds.; Academic Press: Boston, MA, USA, 2014; pp. 245–256. [Google Scholar]
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Khan, F.; Nguyen Pham, D.T.; Oloketuyi, S.F.; Kim, Y.M. Antibiotics and their different application strategies in controlling the biofilm forming pathogenic bacteria. Curr. Pharm. Biotechnol. 2019, 21, 270–286. [Google Scholar] [CrossRef]
- Oloketuyi, S.F.; Khan, F. Strategies for biofilm inhibition and virulence attenuation of foodborne pathogen-Escherichia coli O157:H7. Curr. Microbiol. 2017, 74, 1477–1489. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, J.L.; Patel, R. The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Ther. 2007, 82, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister Cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Bernier, S.; Surette, M. Concentration-dependent activity of antibiotics in natural environments. Front. Microbiol. 2013, 4, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mlynarcik, P.; Kolar, M. Starvation- and antibiotics-induced formation of persister cells in Pseudomonas aeruginosa. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech. Rep. 2017, 161, 58–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Q.; Zhou, W.; Hu, L.; Qi, Y.; Ning, H.; Chen, J.; Mo, H. Bactericidal activity of alpha-bromocinnamaldehyde against persisters in Escherichia coli. PLoS ONE 2017, 12, e0182122. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues Felix, C.; Gupta, R.; Geden, S.; Roberts, J.; Winder, P.; Pomponi, S.A.; Diaz, M.C.; Reed, J.K.; Wright, A.E.; Rohde, K.H. Selective killing of dormant Mycobacterium tuberculosis by marine natural products. Antimicrob. Agents Chemother. 2017, 61, e00743-17. [Google Scholar] [CrossRef] [Green Version]
- Kwan, B.W.; Valenta, J.A.; Benedik, M.J.; Wood, T.K. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 2013, 57, 1468–1473. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-H.; Kim, Y.-G.; Gwon, G.; Wood, T.K.; Lee, J. Halogenated indoles eradicate bacterial persister cells and biofilms. AMB Express 2016, 6, 123. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V.K.; Verma, S.; Gupta, S.; Singh, A.; Pal, A.; Srivastava, S.K.; Srivastava, P.K.; Singh, S.C.; Darokar, M.P. Membrane-damaging potential of natural L-(-)-usnic acid in Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 3375–3383. [Google Scholar] [CrossRef]
- Kwan, B.W.; Chowdhury, N.; Wood, T.K. Combatting bacterial infections by killing persister cells with mitomycin C. Environ. Microbiol. 2015, 17, 4406–4414. [Google Scholar] [CrossRef]
- Hurdle, J.G.; O’Neill, A.J.; Chopra, I.; Lee, R.E. Targeting bacterial membrane function: An underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 2011, 9, 62–75. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, N.; Wood, T.L.; Martínez-Vázquez, M.; García-Contreras, R.; Wood, T.K. DNA-crosslinker cisplatin eradicates bacterial persister cells. Biotechnol. Bioeng. 2016, 113, 1984–1992. [Google Scholar] [CrossRef]
- Conlon, B.P.; Nakayasu, E.S.; Fleck, L.E.; LaFleur, M.D.; Isabella, V.M.; Coleman, K.; Leonard, S.N.; Smith, R.D.; Adkins, J.N.; Lewis, K. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 2013, 503, 365–370. [Google Scholar] [CrossRef] [Green Version]
- Ratti, R.P.; Gomes, B.C.; Martinez, R.C.R.; Souza, V.M.; Martinis, E.C.P.D. Elongated cells of Listeria monocytogenes in biofilms in the presence of sucrose and bacteriocin-producing Leuconostoc mesenteroides A11. Food Sci. Technol. 2010, 30, 1011–1016. [Google Scholar] [CrossRef] [Green Version]
- Zaika, L.L.; Fanelli, J.S. Growth kinetics and cell morphology of Listeria monocytogenes Scott A as affected by temperature, NaCl, and EDTA. J. Food Prot. 2003, 66, 1208–1215. [Google Scholar] [CrossRef]
- Lee, B.-H.; Hébraud, M.; Bernardi, T. Increased adhesion of Listeria monocytogenes strains to abiotic surfaces under cold stress. Front. Microbiol 2017, 8, 2221. [Google Scholar] [CrossRef]
- Rismondo, J.; Halbedel, S.; Gründling, A. Cell shape and antibiotic resistance are maintained by the activity of multiple FtsW and RodA enzymes in Listeria monocytogenes. mBio 2019, 10, e01448-19. [Google Scholar] [CrossRef] [Green Version]
- Van der Veen, S.; van Schalkwijk, S.; Molenaar, D.; de Vos, W.M.; Abee, T.; Wells-Bennik, M.H. The SOS response of Listeria monocytogenes is involved in stress resistance and mutagenesis. Microbiology 2010, 156, 374–384. [Google Scholar] [CrossRef] [Green Version]
- Giesbrecht, P.; Kersten, T.; Maidhof, H.; Wecke, J. Staphylococcal cell wall: Morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 1998, 62, 1371–1414. [Google Scholar] [CrossRef] [Green Version]
- Khan, F.; Manivasagan, P.; Lee, J.W.; Pham, D.T.N.; Oh, J.; Kim, Y.M. Fucoidan-stabilized gold nanoparticle-mediated biofilm inhibition, attenuation of virulence and motility properties in Pseudomonas aeruginosa PAO1. Mar. Drugs 2019, 17, 208. [Google Scholar] [CrossRef] [Green Version]
- Khan, F.; Lee, J.W.; Manivasagan, P.; Pham, D.T.N.; Oh, J.; Kim, Y.M. Synthesis and characterization of chitosan oligosaccharide-capped gold nanoparticles as an effective antibiofilm drug against the Pseudomonas aeruginosa PAO1. Microb. Pathog. 2019, 135, 103623. [Google Scholar] [CrossRef]
- Khan, F.; Manivasagan, P.; Pham, D.T.N.; Oh, J.; Kim, S.K.; Kim, Y.M. Antibiofilm and antivirulence properties of chitosan-polypyrrole nanocomposites to Pseudomonas aeruginosa. Microb. Pathog. 2019, 128, 363–373. [Google Scholar] [CrossRef]









© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, F.; Yu, H.; Kim, Y.-M. Bactericidal Activity of Usnic Acid-Chitosan Nanoparticles against Persister Cells of Biofilm-Forming Pathogenic Bacteria. Mar. Drugs 2020, 18, 270. https://doi.org/10.3390/md18050270
Khan F, Yu H, Kim Y-M. Bactericidal Activity of Usnic Acid-Chitosan Nanoparticles against Persister Cells of Biofilm-Forming Pathogenic Bacteria. Marine Drugs. 2020; 18(5):270. https://doi.org/10.3390/md18050270
Chicago/Turabian StyleKhan, Fazlurrahman, Hongsik Yu, and Young-Mog Kim. 2020. "Bactericidal Activity of Usnic Acid-Chitosan Nanoparticles against Persister Cells of Biofilm-Forming Pathogenic Bacteria" Marine Drugs 18, no. 5: 270. https://doi.org/10.3390/md18050270
APA StyleKhan, F., Yu, H., & Kim, Y. -M. (2020). Bactericidal Activity of Usnic Acid-Chitosan Nanoparticles against Persister Cells of Biofilm-Forming Pathogenic Bacteria. Marine Drugs, 18(5), 270. https://doi.org/10.3390/md18050270