Transcriptomic Analysis of Four Cerianthid (Cnidaria, Ceriantharia) Venoms
Abstract
:1. Introduction
2. Results
2.1. Results for Sequencing and De-Novo Transcriptome Assembly of Four Cerianthid Species
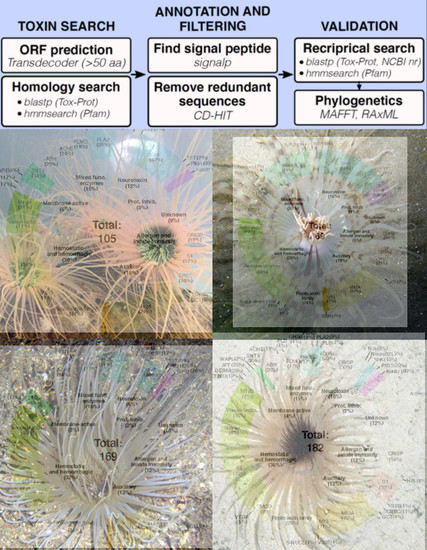
2.2. Diversity and Phylogenetic Analysis of Putative Toxin-Like Gene Profiles for Cerianthids Species
2.2.1. Neurotoxins
2.2.2. Hemostatic and Hemorrhagic Toxins
2.2.3. Membrane-Active Toxins, Protease Inhibitors
2.2.4. Mixed Function Enzymes
2.2.5. Protease Inhibitors
2.2.6. Allergen and Innate Immunity
2.2.7. Venom Auxiliary Proteins
3. Discussion
4. Materials and Methods
4.1. Tissue Collection, RNA Extraction, Next-Gen Sequencing, and Transcriptome Assembly
4.2. Bioinformatic Analysis and Venom Annotation
4.3. Phylogenetic Analysis of Select Gene Families
4.4. Availability of Supporting Data
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cartwright, P.; Collins, A. Fossils and phylogenies: Integrating multiple lines of evidence to investigate the origin of early major metazoan lineages. Integr. Comp. Biol. 2007, 47, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Kayal, E.; Bentlage, B.; Sabrina Pankey, M.; Ohdera, A.H.; Medina, M.; Plachetzki, D.C.; Collins, A.G.; Ryan, J.F. Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol. Biol. 2018, 18, 68. [Google Scholar] [CrossRef] [Green Version]
- Mariscal, R.N. Nematocysts. In Coelenterate Biology: Reviews and New Perspectives; Muscatine, L., Lenhoff, H.M., Eds.; Academic Press: New York, NY, USA, 1974; pp. 129–178. [Google Scholar]
- Fautin, D.G. Structural diversity, systematics, and evolution of cnidae. Toxicon 2009, 54, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Bigger, C.H. Interspecific and intraspecific acrorhagial aggressive behavior among sea anemones: A recognition of self and not-self. Biol. Bull. 1980, 159, 117–134. [Google Scholar] [CrossRef]
- Williams, R.B. Acrorhagi, catch tentacles and sweeper tentacles: A synopsis of ‘aggression’ of actiniarian and scleractinian Cnidaria. In Coelenterate Biology: Recent Research on Cnidaria and Ctenophora; Williams, R.B., Cornelius, P.F.S., Hughes, R.G., Robson, E.A., Eds.; Developments in Hydrobiology Springer: Dordrecht, The Netherlands, 1991; Volume 66, pp. 539–545. [Google Scholar]
- Macrander, J.; Brugler, M.R.; Daly, M. A RNA-seq approach to identify putative toxins from acrorhagi in aggressive and non-aggressive Anthopleura elegantissima polyps. BMC Genom. 2015, 16, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Columbus-Shenkar, Y.Y.; Sachkova, M.Y.; Macrander, J.; Fridrich, A.; Modepalli, V.; Reitzel, A.M.; Sunagar, K.; Moran, Y. Dynamics of venom composition across a complex life cycle. eLife 2018, 7, e35014. [Google Scholar] [CrossRef] [PubMed]
- Schendel, V.; Rash, L.D.; Jenner, R.A.; Undheim, E.A. The diversity of venom: The importance of behavior and venom system morphology in understanding its ecology and evolution. Toxins 2019, 11, 666. [Google Scholar] [CrossRef] [Green Version]
- Turk, T.; Kem, W.R. The phylum Cnidaria and investigations of its toxins and venoms until 1990. Toxicon 2009, 54, 1031–1037. [Google Scholar] [CrossRef]
- Jouiaei, M.; Yanagihara, A.; Madio, B.; Nevalainen, T.; Alewood, P.; Fry, B. Ancient venom systems: A review on Cnidaria toxins. Toxins 2015, 7, 2251–2271. [Google Scholar] [CrossRef] [Green Version]
- Burnett, J.W. Treatment of atlantic cnidarian envenomations. Toxicon 2009, 54, 1201–1205. [Google Scholar] [CrossRef]
- Tibballs, J. Australian venomous jellyfish, envenomation syndromes, toxins and therapy. Toxicon 2006, 48, 830–859. [Google Scholar] [CrossRef] [PubMed]
- Tibballs, J.; Yanagihara, A.A.; Turner, H.C.; Winkel, K. Immunological and toxinological responses to jellyfish stings. Inflamm. Allergy Drug Targets 2011, 10, 438–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badré, S. Bioactive toxins from stinging jellyfish. Toxicon 2014, 91, 114–125. [Google Scholar] [CrossRef]
- Richard, J.; Portier, P. Fascicule 95: Recherches sur la toxine des coelentéres et les phénomènes d’anaphylaxie. In Résultats des Campagnes Scientifiques du Prince de Monaco; Imp. de Monaco: Monaco, 1936. [Google Scholar]
- Lane, C.E.; Dodge, E. The toxicity of Physalia nematocysts. Biol. Bull. 1956, 115, 219–226. [Google Scholar] [CrossRef]
- Tamkun, M.M.; Hessinger, D.A. Isolation and partial characterization of a hemolytic and toxic protein from the nematocyst venom of the Portuguese man-of-war, Physalia physalis. BBA Protein Struct. 1981, 667, 87–98. [Google Scholar] [CrossRef]
- Barnes, J.H. Chironex fleckeri and Chiropsalmus quadrigatus: Morphological distinctions. N. Qld. Nat. 1965, 32, 13–22. [Google Scholar]
- Nagai, H. Recent progress in jellyfish toxin study. J. Health Sci. 2003, 49, 337–340. [Google Scholar] [CrossRef] [Green Version]
- Brinkman, D.L.; Konstantakopoulos, N.; McInerney, B.V.; Mulvenna, J.; Seymour, J.E.; Isbister, G.K.; Hodgson, W.C. Chironex fleckeri (box jellyfish) venom proteins: Expansion of cnidarian toxin family that elicits variable cytolytic and cardiovascular effects. J. Biol. Chem. 2014, 289, 4798–4812. [Google Scholar] [CrossRef] [Green Version]
- Brinkman, D.L.; Jia, X.; Potriquet, J.; Kumar, D.; Dash, D.; Kvaskoff, D.; Mulvenna, J. Transcriptome and venom proteome of the box jellyfish Chironex fleckeri. BMC Genom. 2015, 16, 407. [Google Scholar] [CrossRef] [Green Version]
- Tibballs, J. Australian Chirodropid Cubozoan Jellyfish Envenomation. In Clinical Toxinology in Australia, Europe, and Americas; Vogel, C.W., Seifert, S.A., Tambourgi, D.V., Eds.; Springer: Dordrecht, The Netherlands, 2018; pp. 331–354. [Google Scholar] [CrossRef]
- Goodwin, M.H.; Telford, M. The nematocyst toxin of Metridium. Biol. Bull. 1971, 140, 389–399. [Google Scholar] [CrossRef]
- Madio, B.; King, G.F.; Undheim, E.A.B. Sea anemone toxins: A structural overview. Mar. Drugs 2019, 17, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hessinger, D.A. Nematocyst venoms and toxins. In The Biology of Nematocysts; Hessinger, D.A., Lenhoff, H.M., Eds.; Academic Press: California, CA, USA, 1988; pp. 333–368. [Google Scholar]
- Ashwood, L.M.; Norton, R.S.; Undheim, E.A.B.; Hurwood, D.A.; Prentis, P.J. Characterizing functional venom profiles of anthozoans and medusozoans within their ecological context. Mar. Drugs 2020, 18, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, J.; Peixe, L.; Gomes, N.C.M.; Calado, R. Cnidarians as a Source of New Marine Bioactive Compounds—An Overview of the Last Decade and Future Steps for Bioprospecting. Mar. Drugs 2011, 9, 1860–1886. [Google Scholar] [CrossRef] [PubMed]
- Thangaraj, S.; Bragadeeswaran, S.; Gokula, V. Bioactive compounds of sea anemones: A review. Int. J. Pept. Res. Ther. 2019, 25, 1405–1416. [Google Scholar] [CrossRef]
- Prentis, P.J.; Pavasovic, A.; Norton, R.S. Sea Anemones: Quiet Achievers in the Field of Peptide Toxins. Toxins 2018, 10, 36. [Google Scholar] [CrossRef] [Green Version]
- Castañeda, O.; Sotolongo, V.; Amor, A.M.; Stöcklin, R.; Anderson, A.J.; Harvey, A.L.; Engström, Å.; Wernstedt, C.; Karlsson, E. Characterization of a potassium channel toxin from the Caribbean Sea anemone Stichodactyla helianthus. Toxicon 1995, 33, 603–613. [Google Scholar] [CrossRef]
- Chandy, K.G.; Norton, R.S. Peptide blockers of Kv1. 3 channels in T cells as therapeutics for autoimmune disease. Curr. Opin. Chem. Biol. 2017, 38, 97–107. [Google Scholar] [CrossRef]
- Tarcha, E.J.; Olsen, C.M.; Probst, P.; Peckham, D.; Muñoz-Elías, E.J.; Kruger, J.G.; Iadonato, S.P. Safety and pharmacodynamics of dalazatide, a Kv1. 3 channel inhibitor, in the treatment of plaque psoriasis: A randomized phase 1b trial. PLoS ONE 2017, 12, e0180762. [Google Scholar] [CrossRef]
- Beeton, C.; Pennington, M.W.; Norton, R.S. Analogs of the sea anemone potassium channel blocker ShK for the treatment of autoimmune diseases. Inflamm. Allergy Drug Targets 2011, 10, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Norton, R.S.; Pennington, M.W.; Beeton, C. Case study 2: Transforming a toxin into a therapeutic: The sea anemone potassium channel blocker ShK toxin for treatment of autoimmune diseases. In Venoms to Drugs: Venom as a Source for the Development of Human Therapeutics; King, G.F., Ed.; Royal Society of Chemistry: Cambridge, UK, 2015; pp. 255–274. [Google Scholar] [CrossRef]
- Chen, X.; Leahy, D.; Van Haeften, J.; Hartfield, P.; Prentis, P.J.; van der Burg, C.A.; Surm, J.M.; Pavasovic, A.; Madio, B.; Hamilton, B.R.; et al. A versatile and robust serine protease inhibitor scaffold from Actinia tenebrosa. Mar. Drugs 2019, 17, 701. [Google Scholar] [CrossRef] [Green Version]
- Liao, Q.; Feng, Y.; Yang, B.; Lee, S.M.-Y. Cnidarian peptide neurotoxins: A new source of various ion channel modulators or blockers against central nervous systems disease. Drug Discov. Today 2019, 24, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, H.; Xue, W.; Yue, Y.; Liu, S.; Xing, R.; Li, P. Jellyfish venomics and venom gland transcriptomics analysis of Stomolophus meleagris to reveal the toxins associated with sting. J. Proteom. 2014, 106, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, H.; Yue, Y.; Liu, S.; Xing, R.; Chen, X.; Li, P. Combined proteomics and transcriptomics identifies sting-related toxins of jellyfish Cyanea nozakii. J. Proteom. 2016, 148, 57–64. [Google Scholar] [CrossRef]
- Rachamim, T.; Morgenstern, D.; Aharonovich, D.; Brekhman, V.; Lotan, T.; Sher, D. The dynamically evolving nematocyst content of an anthozoan, a scyphozoan, and a hydrozoan. Mol. Biol. Evol. 2015, 32, 740–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Zhou, Y.; Liu, D.; Wang, Q.; Ruan, Z.; He, Q.; Zhang, L. Global transcriptome analysis of the tentacle of the jellyfish Cyanea capillata using deep sequencing and expressed sequence tags: Insight into the toxin- and degenerative disease-related transcripts. PLoS ONE 2015, 10, e0142680. [Google Scholar] [CrossRef] [Green Version]
- Macrander, J.; Broe, M.; Daly, M. Tissue-specific venom composition and differential gene expression in sea anemones. Genome Biol. Evol. 2016, 8, 2358–2375. [Google Scholar] [CrossRef] [Green Version]
- Ponce, D.; Brinkman, D.; Potriquet, J.; Mulvenna, J. Tentacle transcriptome and venom proteome of the pacific sea nettle, Chrysaora fuscescens (Cnidaria: Scyphozoa). Toxins 2016, 8, 102. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Wang, B.; Wang, B.; Wang, Q.; Liu, G.; Wang, T.; He, Q.; Zhang, L. Unique Diversity of Sting-Related Toxins Based on Transcriptomic and Proteomic Analysis of the Jellyfish Cyanea Capillata and Nemopilema Nomurai (Cnidaria: Scyphozoa). J. Proteome Res. 2018, 18, 436–448. [Google Scholar] [CrossRef]
- Lewis Ames, C.; Ryan, J.F.; Bely, A.E.; Cartwright, P.; Collins, A.G. A New Transcriptome and Transcriptome Profiling of Adult and Larval Tissue in the Box Jellyfish Alatina alata: An Emerging Model for Studying Venom, Vision and Sex. BMC Genom. 2016, 17, 650. [Google Scholar] [CrossRef] [Green Version]
- Madio, B.; Undheim, E.A.B.; King, G.F. Revisiting Venom of the Sea Anemone Stichodactyla haddoni: Omics Techniques Reveal the Complete Toxin Arsenal of a Well-Studied Sea Anemone Genus. J. Proteom. 2017, 166, 83–92. [Google Scholar] [CrossRef]
- Huang, C.; Morlighem, J.-É.R.; Zhou, H.; Lima, É.P.; Gomes, P.B.; Cai, J.; Lou, I.; Pérez, C.D.; Lee, S.M.; Rádis-Baptista, G. The transcriptome of the zoanthid Protopalythoa variabilis (Cnidaria, Anthozoa) predicts a basal repertoire of toxin-like and venom-auxiliary polypeptides. Genome Biol. Evol. 2016, 8, 3045–3064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Q.; Li, S.; Siu, S.W.I.; Yang, B.; Huang, C.; Chan, J.Y.-W.; Morlighem, J.-É.R.L.; Wong, C.T.T.; Rádis-Baptista, G.; Lee, S.M.-Y. Novel kunitz-like peptides discovered in the zoanthid Palythoa caribaeorum through transcriptome sequencing. J. Proteome Res. 2018, 17, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Liao, Q.; Gong, G.; Poon, T.C.W.; Ang, I.L.; Lei, K.M.K.; Siu, S.W.I.; Wong, C.T.T.; Rádis-Baptista, G.; Lee, S.M.-Y. Combined transcriptomic and proteomic analysis reveals a diversity of venom-related and toxin-like peptides expressed in the mat anemone Zoanthus natalensis (Cnidaria, Hexacorallia). Arch. Toxicol. 2019, 93, 1745–1767. [Google Scholar] [CrossRef] [PubMed]
- Rivera-de-Torre, E.; Martínez-del-Pozo, Á.; Garb, J.E. Stichodactyla Helianthus’ de novo transcriptome assembly: Discovery of a new actinoporin isoform. Toxicon 2018, 150, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Surm, J.M.; Smith, H.L.; Madio, B.; Undheim, E.A.B.; King, G.F.; Hamilton, B.R.; Burg, C.A.; Pavasovic, A.; Prentis, P.J. A process of convergent amplification and tissue-specific expression dominates the evolution of toxin and toxin-like genes in sea anemones. Mol. Ecol. 2019, 28, 2272–2289. [Google Scholar] [CrossRef] [Green Version]
- Sachkova, M.Y.; Singer, S.A.; Macrander, J.; Reitzel, A.M.; Peigneur, S.; Tytgat, J.; Moran, Y. The birth and death of toxins with distinct functions: A case study in the sea anemone Nematostella. Mol. Biol. Evol. 2019, 36, 2001–2012. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Carreto, S.; Vera-Estrella, R.; Portillo-Bobadilla, T.; Licea-Navarro, A.; Bernaldez-Sarabia, J.; Rudiño-Piñera, E.; Verleyen, J.J.; Rodríguez, E.; Rodríguez-Almazán, C. Transcriptomic and proteomic analysis of the tentacles and mucus of Anthopleura Dowii Verrill, 1869. Mar. Drugs 2019, 17, 436. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, M.L.; Tonkin-Hill, G.Q.; Morales, R.A.V.; Purcell, A.W.; Papenfuss, A.T.; Norton, R.S. Tentacle transcriptomes of the speckled anemone (Actiniaria: Actiniidae: Oulactis Sp.): Venom-related components and their domain structure. Mar. Biotechnol. 2020, 22, 207–219. [Google Scholar] [CrossRef]
- Zapata, F.; Goetz, F.E.; Smith, S.A.; Howison, M.; Siebert, S.; Church, S.H.; Sanders, S.M.; Ames, C.L.; McFadden, C.S.; France, S.C.; et al. Phylogenomic analyses support traditional relationships within Cnidaria. PLoS ONE 2015, 10, e0139068. [Google Scholar] [CrossRef] [Green Version]
- Chang, E.S.; Neuhof, M.; Rubinstein, N.D.; Diamant, A.; Philippe, H.; Huchon, D.; Cartwright, P. Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc. Natl. Acad. Sci. USA 2015, 112, 14912–14917. [Google Scholar] [CrossRef] [Green Version]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 2012, 60, 551–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, C.; Daly, N.; Wilson, D. Coral venom toxins. Front. Ecol. Evol. 2019, 7, 320. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.; Klug, M.; Tardent, P. Some physical and chemical properties of purified nematocysts of Hydra attenuata Pall. (Hydrozoa, Cnidaria). Comp. Biochem. Physiol. B Biochem. 1987, 88, 855–862. [Google Scholar] [CrossRef]
- Marchini, B.; De, L.N.; Mazzei, M.; Mariottini, G.L. A fast centrifuge method for nematocyst isolation from Pelagia noctiluca Forskal (Cnidaria: Scyphozoa). Riv. Biol. 2004, 97, 505–515. [Google Scholar] [PubMed]
- Carrette, T.; Seymour, J. Cardiotoxic effects of venoms from Chironex fleckeri and Chiropsalmus sp. on an invertebrate model. J. Venom Anim. Toxins 2004, 12, 245–254. [Google Scholar] [CrossRef]
- Feng, J.; Yu, H.; Li, C.; Xing, R.; Liu, S.; Wang, L.; Cai, S.; Li, P. Isolation and characterization of lethal proteins in nematocyst venom of the jellyfish Cyanea nozakii Kishinouye. Toxicon 2010, 55, 118–125. [Google Scholar] [CrossRef]
- Klug, M.; Weber, J.; Tardent, P. Hemolytic and toxic properties of Hydra attenuata nematocysts. Toxicon 1989, 27, 325–339. [Google Scholar] [CrossRef]
- Zenkert, C.; Takahashi, T.; Diesner, M.O.; Özbek, S. Morphological and molecular analysis of the Nematostella vectensis cnidome. PLoS ONE 2011, 6, e22725. [Google Scholar] [CrossRef]
- Von Reumont, B.M. Studying smaller and neglected organisms in modern evolutionary venomics implementing RNASeq (transcriptomics)—A critical guide. Toxins 2018, 10, 292. [Google Scholar] [CrossRef] [Green Version]
- Mariscal, R.N.; Conklin, E.J.; Bigger, C.H. The ptychocyst, a major new category of cnida used in tube construction by a cerianthid anemone. Biol. Bull. 1977, 152, 392–405. [Google Scholar] [CrossRef]
- Stampar, S.N.; Maronna, M.M.; Kitahara, M.V.; Reimer, J.D.; Beneti, J.S.; Morandini, A.C. Ceriantharia in current systematics: Life cycles, morphology and genetics. In The Cnidaria, Past, Present and Future; Goffredo, S., Dubinsky, Z., Eds.; Springer: Cham, Switzerland, 2016; pp. 61–72. [Google Scholar]
- Quattrini, A.M.; Faircloth, B.C.; Dueñas, L.F.; Bridge, T.C.; Brugler, M.R.; Calixto-Botía, I.F.; DeLeo, D.M.; Foret, S.; Herrera, S.; Lee, S.M.; et al. Universal target-enrichment baits for anthozoan (Cnidaria) phylogenomics: New approaches to long-standing problems. Mol. Ecol. Resour. 2018, 18, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.; Brugler, M.R.; Cartwright, P.; Collins, A.G.; Dawson, M.N.; Fautin, D.G.; France, S.C.; Mcfadden, C.S.; Opresko, D.M.; Rodriguez, E.; et al. The phylum Cnidaria: A review of phylogenetic patterns and diversity 300 years after Linnaeus *. Zootaxa 2007, 1668, 127–182. [Google Scholar] [CrossRef] [Green Version]
- Stampar, S.N.; Maronna, M.M.; Kitahara, M.V.; Reimer, J.D.; Morandini, A.C. Fast-evolving mitochondrial DNA in Ceriantharia: A reflection of Hexacorallia paraphyly? PLoS ONE 2014, 9, e86612. [Google Scholar] [CrossRef] [PubMed]
- Stampar, S.N.; Broe, M.B.; Macrander, J.; Reitzel, A.M.; Brugler, M.R.; Daly, M. Linear mitochondrial genome in Anthozoa (Cnidaria): A case study in Ceriantharia. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Bridge, D.; Cunningham, C.W.; DeSalle, R.; Buss, L.W. Class-level relationships in the phylum Cnidaria: Molecular and morphological evidence. Mol. Biol. Evol. 1995, 12, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Kayal, E.; Roure, B.; Philippe, H.; Collins, A.G.; Lavrow, D.V. Cnidarian phylogenetic relationships as revealed by mitogenomics. BMC Evol. Biol. 2013, 13, 5. [Google Scholar] [CrossRef] [Green Version]
- Stampar, S.N.; Morandini, A.C.; Branco, L.C.; Da Silveira, F.L.; Migotto, A.E. Drfting in the oceans: Isarachnanthus nocturnus (Cnidaria, Ceriantharia, Arachnactidae), an anthozoan with an extended planktonic stage. Mar. Biol. 2015, 162, 2161–2169. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [Green Version]
- Bateman, A. The Pfam Protein Families Database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Griselda, A.V. Molecular Characterization of Carukia Barnesi and Malo Kingi, Cnidaria; Cubozoa; Carybdeidae. Doctoral Dissertation, James Cook University, Queensland, Australia, 2009. Research Online at JCU. Available online: https://researchonline.jcu.edu.au/8218/ (accessed on 5 April 2020).
- Rangaraju, S.; Khoo, K.K.; Feng, Z.P.; Crossley, G.; Nugent, D.; Khaytin, I.; Chi, V.; Pham, C.; Calabresi, P.; Pennington, M.W.; et al. Potassium channel modulation by a toxin domain in matrix metalloprotease 23. J. Biol. Chem. 2010, 285, 9124–9136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krebs, H.C.; Habermehl, G.G. Isolation and structural determination of a hemolytic active peptide from the sea anemone Metridium senile. Naturwissenschaften 1987, 74, 395–396. [Google Scholar] [CrossRef]
- von Reumont, B.M.; Campbell, L.I.; Richter, S.; Hering, L.; Sykes, D.; Hetmank, J.; Jenner, R.A.; Bleidorn, C. A polychaete’s powerful punch: Venom gland transcriptomics of Glycera reveals a complex cocktail of toxin homologs. Genome Biol. Evol. 2014, 6, 2406–2423. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Jiang, G.; Wang, T.; Zhang, J.; Liu, W.; Xu, Z.; Zhang, J.; Xiao, L. An integrated transcriptomic and proteomic analysis reveals toxin arsenal of a novel antarctic jellyfish Cyanea Sp. J. Proteom. 2019, 208, 103483. [Google Scholar] [CrossRef] [PubMed]
- Jaimes-Becerra, A.; Gacesa, R.; Doonan, L.B.; Hartigan, A.; Marques, A.C.; Okamura, B.; Long, P.F. “Beyond primary sequence”—Proteomic data reveal complex toxins in cnidarian venoms. Integr. Comp. Biol. 2019, 59, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, M.B.; Flores-Torres, A.; Batista, C.V.; Falcón, A.; López-Vera, E.; de la Cotera, E.P. Structural characterization of five post-translationally modified isomorphs of a novel putative delta-conotoxin from the vermivorous snail Conus delessertii from the Mexican Caribbean sea. Peptides 2009, 30, 458–466. [Google Scholar] [CrossRef]
- Gonzales, D.T.T.; Saloma, C.P. A bioinformatics survey for conotoxin-like sequences in three turrid snail venom duct transcriptomes. Toxicon 2014, 92, 66–74. [Google Scholar] [CrossRef]
- Kini, R.M. Evolution of three-finger toxins—A versatile mini protein scaffold. Acta Chim. Slov. 2011, 58, 693–701. [Google Scholar]
- Kuhn, P.; Deacon, A.M.; Comoso, S.; Rajaseger, G.; Kini, R.M.; Usón, I.; Kolatkar, P.R. The atomic resolution structure of bucandin, a novel toxin isolated from the malayan krait, determined by direct methods. Acta Cryst. D Biol. Cryst. 2000, 56, 1401–1407. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.A.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [Green Version]
- Arlinghaus, F.T.; Eble, J.A. C-Type lectin-like proteins from snake venoms. Toxicon 2012, 60, 512–519. [Google Scholar] [CrossRef]
- OmPraba, G.; Chapeaurouge, A.; Doley, R.; Devi, K.R.; Padmanaban, P.; Venkatraman, C.; Velmurugan, D.; Lin, Q.; Kini, R.M. Identification of a novel family of snake venom proteins veficolins from Cerberus rynchops using a venom gland transcriptomics and proteomics approach. J. Proteome Res. 2010, 9, 1882–1893. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M. The intriguing world of prothrombin activators from snake venom. Toxicon 2005, 45, 1133–1145. [Google Scholar] [CrossRef]
- Balasubramanian, P.G.; Beckmann, A.; Warnken, U.; Schnölzer, M.; Schüler, A.; Bornberg-Bauer, E.; Holstein, T.W.; Özbek, S. Proteome of Hydra nematocyst. J. Biol. Chem. 2012, 287, 9672–9681. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Anderluh, G.; Maček, P. Dissecting the actinoporin pore-forming mechanism. Structure 2003, 11, 1312–1313. [Google Scholar] [CrossRef] [Green Version]
- Violette, A.; Biass, D.; Dutertre, S.; Koua, D.; Piquemal, D.; Pierrat, F.; Stöcklin, R.; Favreau, P. Large-scale discovery of conopeptides and conoproteins in the injectable venom of a fish-hunting cone snail using a combined proteomic and transcriptomic approach. J. Proteom. 2012, 75, 5215–5225. [Google Scholar] [CrossRef]
- Warren, W.C.; Hillier, L.W.; Graves, J.A.M.; Birney, E.; Ponting, C.P.; Grützner, F.; Belov, K.; Miller, W.; Clarke, L.; Chinwalla, A.T.; et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature 2008, 453, 175–183. [Google Scholar] [CrossRef] [PubMed]
- García-Ortega, L.; Alegre-Cebollada, J.; García-Linares, S.; Bruix, M.; Martínez-del-Pozo, Á.; Gavilanes, J.G. The behavior of sea anemone actinoporins at the water–membrane interface. BBA Biomembr. 2011, 1808, 2275–2288. [Google Scholar] [CrossRef] [Green Version]
- Andrich, F.; Carnielli, J.B.T.; Cassoli, J.S.; Lautner, R.Q.; Santos, R.A.S.; Pimenta, A.M.C.; de Lima, M.E.; Figueiredo, S.G. A potent vasoactive cytolysin isolated from Scorpaena Plumieri scorpionfish venom. Toxicon 2010, 56, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Kiriake, A.; Suzuki, Y.; Nagashima, Y.; Shiomi, K. Proteinaceous toxins from three species of scorpaeniform fish (Lionfish Pterois Lunulata, Devil Stinger Inimicus Japonicus and Waspfish Hypodytes Rubripinnis): Close similarity in properties and primary structures to stonefish toxins. Toxicon 2013, 70, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Whittington, C.M.; Papenfuss, A.T.; Locke, D.P.; Mardis, E.R.; Wilson, R.K.; Abubucker, S.; Mitreva, M.; Wong, E.S.; Hsu, A.L.; Kuchel, P.W.; et al. Novel venom gene discovery in the platypus. Genome Biol. 2010, 11, R95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, A.; Suzuki, M.; Honma, T.; Nagai, H.; Nagashima, Y.; Shiomi, K. Purification, properties and cdna cloning of neoverrucotoxin (NeoVTX), a hemolytic lethal factor from the stonefish Synanceia Verrucosa venom. BBA Gen. Subj. 2006, 1760, 1713–1722. [Google Scholar] [CrossRef]
- Yazawa, K.; Wang, J.-W.; Hao, L.-Y.; Onoue, Y.; Kameyama, M. Verrucotoxin, a stonefish venom, modulates calcium channel activity in guinea-pig ventricular myocytes: Verrucotoxin modulates cardiac calcium channels. Br. J. Pharmacol. 2009, 151, 1198–1203. [Google Scholar] [CrossRef] [Green Version]
- Nair, D.G.; Fry, B.G.; Alewood, P.; Kumar, P.P.; Kini, R.M. Antimicrobial activity of omwaprin, a new member of the waprin family of snake venom proteins. Biochem. J. 2007, 402, 93–104. [Google Scholar] [CrossRef]
- Liu, D.; Wang, Y.; Wei, L.; Ye, H.; Liu, H.; Wang, L.; Liu, R.; Li, D.; Lai, R. Snake venom-like waprin from the frog of Ceratophrys calcarata contains antimicrobial function. Gene 2013, 514, 99–104. [Google Scholar] [CrossRef]
- Hessinger, D.A.; Lenhoff, H.M. Mechanism of Hemolysis Induced by Nematocyst Venom: Roles of Phospholipase A and Direct Lytic Factor. Arch. Biochem. Biophys. 1976, 173, 603–613. [Google Scholar] [CrossRef]
- Kini, R.M. Excitement ahead: Structure, function and mechanism of snake venom phospholipase a2 enzymes. Toxicon 2003, 42, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Nevalainen, T.J.; Peuravuori, H.J.; Quinn, R.J.; Llewellyn, L.E.; Benzie, J.A.H.; Fenner, P.J.; Winkel, K.D. Phospholipase A2 in Cnidaria. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2004, 139, 731–735. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Ghomashchi, F.; Gelb, M.H.; Dooley, D.J.; Stoehr, S.J.; Giordani, A.B.; Naisbitt, S.R.; Olivera, B.M. Conodipine-M, a novel phospholipase A2 isolated from the venom of the marine snail Conus Magus. J. Biol. Chem. 1995, 270, 3518–3526. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Lynch, K.R. Brown recluse spider (Loxosceles Reclusa) venom phospholipase D (PLD) generates lysophosphatidic acid (LPA). Biochem. J. 2005, 391, 317–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, H.; Lee, K.S.; Kim, B.Y.; Zou, F.M.; Yoon, H.J.; Je, Y.H.; Li, J.; Jin, B.R. A spider-derived kunitz-type serine protease inhibitor that acts as a plasmin inhibitor and an elastase inhibitor. PLoS ONE 2013, 8, e53343. [Google Scholar] [CrossRef]
- Mihelič, M.; Turk, D. Two decades of thyroglobulin type-1 domain research. Biol. Chem. 2007, 388, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Horiike, T.; Nagai, H.; Kitani, S. Identification of allergens in the box jellyfish Chironex Yamaguchii that cause sting dermatitis. Int. Arch. Allergy Immunol. 2015, 167, 73–82. [Google Scholar] [CrossRef]
- Gibbs, G.M.; Roelants, K.; O’Bryan, M.K. The CAP superfamily: Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins—Roles in reproduction, cancer, and immune defense. Endocr. Rev. 2008, 29, 865–897. [Google Scholar] [CrossRef]
- Verdes, A.; Simpson, D.; Holford, M. Are fireworms venomous? Evidence for the convergent evolution of toxin homologs in three species of fireworms (Annelida, Amphinomidae). Genome Biol. Evol. 2018, 10, 249–268. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, Y.; Koike, H.; Sugiyama, Y.; Motoyoshi, K.; Wada, T.; Hishinuma, S.; Mita, M.; Morita, T. Cloning and characterization of novel snake venom proteins that block smooth muscle contraction: Novel proteins in snake venoms. Eur. J. Biochem. 2002, 269, 2708–2715. [Google Scholar] [CrossRef]
- Milne, T.J.; Abbenante, G.; Tyndall, J.D.A.; Halliday, J.; Lewis, R.J. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J. Biol. Chem. 2003, 278, 31105–31110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, K.S.; Vitale, M.; Fehlner, P.; King, T.P. CDNA cloning and primary structure of a white-face hornet venom allergen, antigen 5. Proc. Natl. Acad. Sci. USA 1988, 85, 895–899. [Google Scholar] [CrossRef] [Green Version]
- Ruiming, Z.; Yibao, M.; Yawen, H.; Zhiyong, D.; Yingliang, W.; Zhijian, C.; Wenxin, L. Comparative venom gland transcriptome analysis of the scorpion Lychas mucronatus reveals intraspecific toxic gene diversity and new venomous components. BMC Genom. 2010, 11, 452. [Google Scholar] [CrossRef] [Green Version]
- Ben-Ari, H.; Paz, M.; Sher, D. The chemical armament of reef-building corals: Inter- and intra-specific variation and the identification of an unusual actinoporin in Stylophora Pistilata. Sci. Rep. 2018, 8, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Jung, E.; Kang, C.; Yoon, W.D.; Kim, J.-S.; Kim, E. Scyphozoan jellyfish venom metalloproteinases and their role in the cytotoxicity. Toxicon 2011, 58, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Praher, D.; Schlesinger, A.; Ayalon, A.; Tal, Y.; Technau, U. Analysis of soluble protein contents from the nematocysts of a model sea anemone sheds light on venom evolution. Mar. Biotechnol. 2013, 15, 329–339. [Google Scholar] [CrossRef] [Green Version]
- Fox, J.W.; Serrano, S.M.T. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon 2005, 45, 969–985. [Google Scholar] [CrossRef]
- Weston, A.J.; Chung, R.; Dunlap, W.C.; Morandini, A.C.; Marques, A.C.; Moura-da-Silva, A.M.; Ward, M.; Padilla, G.; da Silva, L.F.; Andreakis, N.; et al. Proteomic characterization of toxins isolated from nematocysts of the south atlantic jellyfish Olindias Sambaquiensis. Toxicon 2013, 71, 11–17. [Google Scholar] [CrossRef]
- Podobnik, M.; Anderluh, G. Pore-forming toxins in Cnidaria. Semin. Cell Dev. Biol. 2017, 72, 133–141. [Google Scholar] [CrossRef]
- Carrette, T.; Alderslade, P.; Seymour, J. Nematocyst ratio and prey in two australian cubomedusans, Chironex fleckeri and Chiropsalmus Sp. Toxicon 2002, 40, 1547–1551. [Google Scholar] [CrossRef]
- Courtney, R.; Sachlikidis, N.; Jones, R.; Seymour, J. Prey capture ecology of the cubozoan Carukia barnesi. PLoS ONE 2015, 10, e0124256. [Google Scholar] [CrossRef] [PubMed]
- Gacesa, R.; Chung, R.; Dunn, S.R.; Weston, A.J.; Jaimes-Becerra, A.; Marques, A.C.; Morandini, A.C.; Hranueli, D.; Starcevic, A.; Ward, M.; et al. Gene duplications are extensive and contribute significantly to the toxic proteome of nematocysts isolated from Acropora Digitifera (Cnidaria: Anthozoa: Scleractinia). BMC Genom. 2015, 16, 774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frazão, B.; Vasconcelos, V.; Antunes, A. Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: An overview. Mar. Drugs 2012, 10, 1812–1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jouiaei, M.; Sunagar, K.; Federman Gross, A.; Scheib, H.; Alewood, P.F.; Moran, Y.; Fry, B.G. Evolution of an ancient venom: Recognition of a novel family of cnidarian toxins and the common evolutionary origin of sodium and potassium neurotoxins in sea anemone. Mol. Biol. Evol. 2015, 32, 1598–1610. [Google Scholar] [CrossRef] [Green Version]
- Sunagawa, S.; DeSalvo, M.K.; Voolstra, C.R.; Reyes-Bermudez, A.; Medina, M. Identification and gene expression analysis of a taxonomically restricted cysteine-rich protein family in reef-building corals. PLoS ONE 2009, 4, e4865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafee, T.; Mitchell, M.L.; Norton, R.S. Mapping the chemical and sequence space of the ShKT superfamily. Toxicon 2019, 165, 95–102. [Google Scholar] [CrossRef]
- Surm, J.M.; Stewart, Z.K.; Papanicolaou, A.; Pavasovic, A.; Prentis, P.J. The draft genome of Actinia Tenebrosa reveals insights into toxin evolution. Ecol. Evol. 2019, 9, 11314–11328. [Google Scholar] [CrossRef] [Green Version]
- Niermann, C.N.; Tate, T.G.; Suto, A.L.; Barajas, R.; White, H.A.; Guswiler, O.D.; Secor, S.M.; Rowe, A.H.; Rowe, M.P. Defensive venoms: Is pain sufficient for predator deterrence? Toxins 2020, 12, 260. [Google Scholar] [CrossRef] [Green Version]
- Sunagar, K.; Morgenstern, D.; Reitzel, A.M.; Moran, Y. Ecological venomics: How genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J. Proteom. 2016, 135, 62–72. [Google Scholar] [CrossRef]
- Arbuckle, K. From Molecules to macroevolution: Venom as a model system for evolutionary biology across levels of life. Toxicon X 2020, 6, 100034. [Google Scholar] [CrossRef]
- Stampar, S.N.; El Didi, S.O.; Paulay, G.; Berumen, M.L. A new species of Arachnanthus from the Red Sea (Cnidaria, Ceriantharia). Zookeys 2018, 748, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haddock, S.H.; Dunn, C.W. Fluorescent proteins function as a prey attractant: Experimental evidence from the hydromedusa Olindias formosus and other marine organisms. Biol. Open 2015, 4, 1094–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macrander, J.; Broe, M.; Daly, M. Multi-copy venom genes hidden in de novo transcriptome assemblies, a cautionary tale with the snakelocks sea anemone Anemonia sulcata (Pennant, 1977). Toxicon 2015, 108, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Holding, M.; Margres, M.; Mason, A.; Parkinson, C.; Rokyta, D. Evaluating the performance of de novo assembly methods for venom-gland transcriptomics. Toxins 2018, 10, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariottini, G.; Pane, L. Cytotoxic and cytolytic cnidarian venoms. A review on health implications and possible therapeutic applications. Toxins 2013, 6, 108. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 17 November 2016).
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from rna-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Altschul, S. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [Green Version]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Godzik, A. Cd-Hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 12 December 2019).
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2016; Available online: http://www.rstudio.com/ (accessed on 5 September 2017).
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [Green Version]







| Species | Reads (PE) | Transcripts | Genes | N50 | BUSCO % |
|---|---|---|---|---|---|
| C. brasiliensis | 34,877,883 | 131,550 | 110,524 | 1276 | 95.4% |
| I. nocturnus | 31,028,274 | 92,757 | 78,821 | 1170 | 89.2% |
| P. borealis | 36,520,791 | 158,633 | 120,542 | 1282 | 97.9% |
| P. maua. | 27,865,720 | 179,576 | 145,788 | 1101 | 88.1% |
| Toxin Family ID | Pfam Domain | Cebr | Isn | Pasb | Pasm |
|---|---|---|---|---|---|
| Neurotoxin (%) | - | 7.1 | 17.4 | 11.0 | 13.3 |
| 332-1 propeptide toxin | ShK | 0 | 0 | 1 | 0 |
| Cysteine-rich venom protein | CAP | 2 | 1 | 10 | 2 |
| ShK-domain | ShK | 6 | 10 | 3 | 8 |
| Three-finger toxin | / | 1 | 1 | 0 | 1 |
| Turripeptide | Kazal_1 | 3 | 0 | 5 | 2 |
| U-actitoxin-Avd9a | ShK | 0 | 0 | 0 | 1 |
| U33-theraphotoxin-Cg1b | / | 0 | 0 | 1 | 0 |
| Hemostatic and Hemorrhagic Toxin (%) | - | 37.3 | 30.4 | 41.2 | 33.3 |
| Beta-fibrinogenase mucrofibrase-3 | Trysin | 0 | 0 | 0 | 1 |
| Blarina Toxin | Trysin | 3 | 0 | 1 | 0 |
| C-type lectin lectoxin | Lectin_C | 6 | 2 | 3 | 1 |
| Coagualtion factor X | Trypin | 1 | 2 | 2 | 0 |
| Coagulation factor V | F5_F8_type_C | 2 | 1 | 6 | 3 |
| Coagulation factor X-activating enzyme heavy chain | Pep_M12B_propep/Reprolysin | 1 | 0 | 1 | 0 |
| Galactose-specific lectin | Lectin_C | 4 | 0 | 9 | 3 |
| Ryncolin | Fibrinogen_C | 8 | 3 | 8 | 6 |
| Snaclec | Lectin_C | 2 | 1 | 3 | 0 |
| Snake venom 5′-nucleotidase | 5_nucleotid_C | 1 | 0 | 1 | 0 |
| Snake venom serine proteinase | Trypsin | 0 | 0 | 0 | 1 |
| Snake venom VEGF | PDGF | 0 | 1 | 1 | 0 |
| Thrombin-like enzyme | Trypsin | 1 | 0 | 3 | 0 |
| Thyrostimulin | DAN | 1 | 0 | 0 | 0 |
| Veficolin-1 | Collagen | 14 | 2 | 9 | 5 |
| Venom peptide isomerase heavy chain | Trypsin | 2 | 0 | 1 | 0 |
| Venom prothrombin activator (F5/F8 type C) | F5_F8_type_C | 6 | 3 | 15 | 4 |
| Venom prothrombin activator (Trypsin) | Trypsin | 9 | 5 | 8 | 7 |
| Zinc metalloproteinase-disintegrin | Pep_M12B_propep/Reprolysin | 2 | 1 | 4 | 4 |
| Membrane-Active (%) | - | 3.6 | 0 | 4.4 | 5.7 |
| DELTA-thalatoxin-Avl2a | MAPF | 0 | 0 | 1 | 1 |
| Jellyfish Toxin | / | 1 | 0 | 2 | 1 |
| Millepora cytotoxin | DERM | 0 | 0 | 2 | 0 |
| Stonutoxin/Neoverrucotoxin | / | 5 | 0 | 2 | 2 |
| Waprin | WAP | 0 | 0 | 1 | 2 |
| Mixed function enzyme (%) | - | 20.7 | 21.7 | 12.6 | 16.2 |
| Acetylcholinesterase | COesterase | 5 | 2 | 3 | 3 |
| Gilatoxin | Trypsin | 0 | 1 | 0 | 0 |
| L-amino-acid oxidase | Amino_oxidase | 6 | 1 | 4 | 3 |
| Peroxiredoxin | AhpC-TSA | 0 | 0 | 1 | 1 |
| Phospholipase-A2/Conodpine | Phospholip_A2 | 5 | 6 | 2 | 5 |
| Phospholipase-B | Phospholip_B | 1 | 1 | 1 | 0 |
| Phospholipase-D | / | 4 | 0 | 1 | 0 |
| Putative endothelial lipase | Lipase | 5 | 1 | 3 | 2 |
| Putative lysosomal acid lipase/cholesteryl ester hydrolase | Abhydro_lipase/Abhydrolase_1 | 4 | 3 | 3 | 3 |
| Trehalase | Trehalase | 0 | 0 | 1 | 0 |
| Venom phosphodiesterase | Phosphodiest | 5 | 0 | 4 | 0 |
| Protease Inhibitor (%) | - | 2.4 | 4.3 | 2.7 | 2.9 |
| Kunitz-type serine protease inhibitor | Knuitz_BPTI | 3 | 3 | 4 | 1 |
| U-actitoxin-Avd3m | Knuitz_BPTI | 0 | 0 | 0 | 1 |
| U24-ctenitoxin-Pn1a | Thyroglobin_1 | 1 | 0 | 1 | 1 |
| Allergen and Innate Immunity (%) | - | 12.7 | 2.2 | 13.2 | 5.4 |
| CRISP/Allergen/PR-1 | CAP | 1 | 0 | 1 | 0 |
| Venom allergen | CAP | 14 | 2 | 12 | 3 |
| Venom phosphatase | His_Phos_2 | 1 | 1 | 1 | 0 |
| Venom protease | Trysin | 1 | 0 | 3 | 3 |
| Venom serine carboxypeptidase | Peptidase_S10 | 0 | 0 | 1 | 0 |
| Venom serine protease | Trysin | 6 | 1 | 6 | 3 |
| Techylectin-like | Fibrinogen_C | 1 | 0 | 0 | 1 |
| Auxiliary Protein (%) | - | 14.8 | 20.2 | 14.8 | 19.0 |
| Astacin-like metalloprotease toxin | Astacin | 6 | 5 | 8 | 9 |
| Cystatin | Cystatin | 0 | 0 | 0 | 1 |
| Glutaminyl-peptide cyclotransferase | Peptidase_M28 | 1 | 1 | 1 | 1 |
| Hyaluronidase | Glyco_hydro_56 | 4 | 0 | 0 | 0 |
| Nematocyst expressed protein | Astacin | 6 | 3 | 11 | 6 |
| Neprilysin | Peptidase_M13_N | 1 | 1 | 3 | 0 |
| Reticulocalbin | EF-hand_7 | 5 | 3 | 3 | 2 |
| Venom protein 302 | IGFBP | 2 | 1 | 1 | 1 |
| TOTAL | - | 169 | 69 | 182 | 105 |
| Unknown | - | 25 | 5 | 24 | 11 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klompen, A.M.L.; Macrander, J.; Reitzel, A.M.; Stampar, S.N. Transcriptomic Analysis of Four Cerianthid (Cnidaria, Ceriantharia) Venoms. Mar. Drugs 2020, 18, 413. https://doi.org/10.3390/md18080413
Klompen AML, Macrander J, Reitzel AM, Stampar SN. Transcriptomic Analysis of Four Cerianthid (Cnidaria, Ceriantharia) Venoms. Marine Drugs. 2020; 18(8):413. https://doi.org/10.3390/md18080413
Chicago/Turabian StyleKlompen, Anna M. L., Jason Macrander, Adam M. Reitzel, and Sérgio N. Stampar. 2020. "Transcriptomic Analysis of Four Cerianthid (Cnidaria, Ceriantharia) Venoms" Marine Drugs 18, no. 8: 413. https://doi.org/10.3390/md18080413
APA StyleKlompen, A. M. L., Macrander, J., Reitzel, A. M., & Stampar, S. N. (2020). Transcriptomic Analysis of Four Cerianthid (Cnidaria, Ceriantharia) Venoms. Marine Drugs, 18(8), 413. https://doi.org/10.3390/md18080413