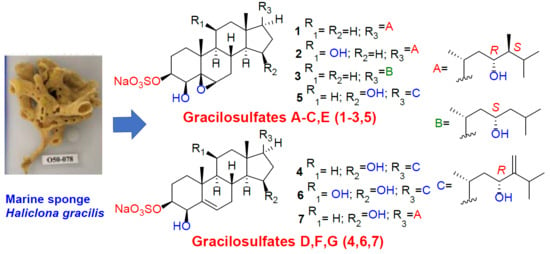
Gracilosulfates A–G, Monosulfated Polyoxygenated Steroids from the Marine Sponge Haliclona gracilis
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Procedures
3.2. Animal Material
3.3. Extraction and Isolation
3.4. Compound Characterization Data
Preparation of MTPA esters of compounds 1, 3, and 4
3.5. Bioactivity Assay
3.5.1. Reagents
3.5.2. Cell Lines and Culture Conditions
3.5.3. In Vitro MTT-Based Drug Sensitivity Assay
3.5.4. Western Blotting
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine Natural Products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef]
- Carvalhal, F.; Correia-da-Silva, M.; Sousa, E.; Pinto, M.; Kijjoa, A. Sources and biological activities of marine sulfated steroids. J. Mol. Endocrinol. 2018, 61, T211–T231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornprobst, J.M.; Sallenave, C.; Barnathan, G. Sulfated compounds from marine organisms. Comp. Biochem. Physiol. 1998, 119B, 1–51. [Google Scholar]
- Kellner Filho, L.C.; Picao, B.W.; Silva Marcio, L.A.; Cunha, W.R.; Pauletti, P.M.; Dias, G.M.; Copp, B.R.; Bertanha, C.S.; Januario, A.H. Bioactive aliphatic sulfates from marine invertebrates. Mar. Drugs 2019, 17, 527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.-M.; Marron, M.T.; Seddon, E.; McLaughlin, S.P.; Ray, D.T.; Whitesell, L.; Gunatilaka, A.A.L. 2,3-Dihydrowithaferin A-3β-O-sulfate, a new potential prodrug of withaferin A from aeroponically grown Withania somnifera. Bioorg. Med. Chem. 2009, 17, 2210–2214. [Google Scholar] [CrossRef]
- Morinaka, B.I.; Pawlik, J.R.; Molinski, T.F. Amaranzoles B-F, imidazole-2-carboxy steroids from the marine sponge Phorbasam aranthus. C24-N- and C24-O-analogues from a divergent oxidative biosynthesis. J. Org. Chem. 2010, 75, 2453–2460. [Google Scholar] [CrossRef] [Green Version]
- Guzii, A.G.; Makarieva, T.N.; Denisenko, V.A.; Dmitrenok, P.S.; Burtseva, Y.V.; Krasokhin, V.B.; Stonik, V.A. Topsentiasterol sulfates with novel iodinated and chlorinated side chains from a marine sponge Topsentia sp. Tetrahedron Lett. 2008, 49, 7191–7193. [Google Scholar] [CrossRef]
- Tabakmakher, K.M.; Makarieva, T.N.; Denisenko, V.A.; Popov, R.S.; Dmitrenok, P.S.; Dyshlovoy, S.A.; Grebnev, B.B.; Bokemeyer, C.; von Amsberg, G.; Cuong, N.X. New trisulfated steroids from the vietnamese marine sponge Halichondria vansoesti and their PSA expression and glucose uptake inhibitory activities. Mar. Drugs 2019, 17, 445. [Google Scholar] [CrossRef] [Green Version]
- Woo, J.-K.; Ha, T.K.Q.; Oh, D.-C.; Oh, W.-K.; Oh, K.-B.; Shin, J. Polyoxygenated steroids from the sponge Clathria gombawuiensis. J. Nat. Prod. 2017, 80, 3224–3233. [Google Scholar] [CrossRef]
- Boonlarppradab, C.; Faulkner, D.J. Eurysterols A and B, cytotoxic and antifungal steroidal sulfates from a marine sponge of the genus Euryspongia. J. Nat. Prod. 2007, 70, 846–848. [Google Scholar] [CrossRef]
- Zhang, H.J.; Sun, J.B.; Lin, H.W.; Wang, Z.L.; Tang, H.; Cheng, P.; Chen, W.S.; Yi, Y.H. A new cytotoxic cholesterol sulfate from marine sponge Halichondria rugosa. Nat. Prod. Res. 2007, 21, 953–958. [Google Scholar] [CrossRef] [PubMed]
- Makarieva, T.N.; Stonik, V.A.; D’yachuk, O.G.; Dmitrenok, A.S. Annasterol sulfate, a novel marine sulfated steroid, inhibitor of glucanase activity from the deep-water sponge Poecillastra laminaris. Tetrahedron Lett. 1995, 36, 129–132. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Matsunaga, S.; Fusetani, N.; Van Soest, R.W.M. Acanthosterol sulfates A−J: Ten new antifungal steroidal sulfates from a marine sponge Acanthodendrilla sp. J. Nat. Prod. 1998, 61, 1374–1378. [Google Scholar] [CrossRef] [PubMed]
- Gabant, M.; Schmitz-Afonso, I.; Gallard, J.-F.; Menou, J.-L.; Laurent, D.; Debitus, C.; Al-Mourabit, A. Sulfated steroids: Ptilosteroids A−C and ptilosaponosides A and B from the Solomon Islands marine sponge Ptilocaulis spiculifer. J. Nat. Prod. 2009, 72, 760–763. [Google Scholar] [CrossRef]
- DiGirolamo, J.A.; Li, X.-C.; Jacob, M.R.; Clark, A.M.; Ferreira, D. Reversal of fluconazole resistance by sulfated sterols from the marine sponge Topsentia sp. J. Nat. Prod. 2009, 72, 1524–1528. [Google Scholar] [CrossRef] [Green Version]
- Radwan, M.M.; Manly, S.P.; Ross, S.A. Two new sulfated sterols from the marine sponge Lendenfeldia dendyi. Nat. Prod. Commun. 2007, 2, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Sperry, S.; Crews, P. Haliclostanone sulfate and halistanol sulfate from an Indo-Pacific Haliclona sponge. J. Nat. Prod. 1997, 60, 29–32. [Google Scholar] [CrossRef]
- Fusetani, N.; Matsunaga, S.; Konosu, S. Bioactive marine metabolites II. Halistanol sulfate, an antimicrobial novel steroid sulfate from the marine sponge halichondria cf. moorei bergquist. Tetrahedron Lett. 1981, 22, 1985–1988. [Google Scholar] [CrossRef]
- Cheng, J.-F.; Lee, J.-S.; Sun, F.; Jares-Erijman, E.A.; Cross, S.; Rinehart, K.L. Hamigerols A and B, unprecedented polysulfate sterol dimers from the mediterranean sponge Hamigera hamigera. J. Nat. Prod. 2007, 70, 1195–1199. [Google Scholar] [CrossRef]
- Ushiyama, S.; Umaoka, H.; Kato, H.; Suwa, Y.; Morioka, H.; Rotinsulu, H.; Losung, F.; Mangindaan, R.E.P.; de Voogd, N.J.; Yokosawa, H.; et al. Manadosterols A and B, sulfonated sterol dimers inhibiting the ubc13-uev1a interaction, isolated from the marine sponge Lissodendryx fibrosa. Nat. Prod. 2012, 75, 1495–1499. [Google Scholar] [CrossRef]
- Guzii, A.G.; Makarieva, T.N.; Denisenko, V.A.; Gerasimenko, A.V.; Udovenko, A.A.; Popov, R.S.; Dmitrenok, P.S.; Golotin, V.A.; Fedorov, S.N.; Grebnev, B.B.; et al. Guitarrins A-E and Aluminumguitarrin A: 5-azaindoles from the northwestern pacific marine sponge Guitarra fimbriata. J. Nat. Prod. 2019, 82, 1704–1709. [Google Scholar] [CrossRef] [PubMed]
- Shubina, L.K.; Makarieva, T.N.; von Amsberg, G.; Denisenko, V.A.; Popov, R.S.; Dmitrenok, P.S.; Dyshlovoy, S.A. Monanchoxymycalin C with anticancer properties, new analogue of crambescidin 800 from the marine sponge Monanchora pulchra. Nat. Prod. Res. 2019, 33, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- Appeltans, W.; Boucher, P.; Boxshall, G.A.; De Broyer, C.; de Voogd, N.J.; Gordon, D.P.; Hoeksema, B.W.; Horton, T.; Kennedy, M.; Mees, J.; et al. (Eds.) World Register of Marine Species. 2012. Available online: http://www.marinespecies.org (accessed on 20 November 2019).
- Santafe´, G.; Paz, V.; Rodrıguez, J.; Jimenez, C. Novel cytotoxic oxygenated C29 sterols from the Colombian marine sponge Polymastia tenax. J. Nat. Prod. 2002, 65, 1161–1164. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xi, Y.; Huang, L.; Li, G.; Mao, Q.; Fang, C.; Shan, T.; Jiang, W.; Zhao, M.; He, W.; et al. A Steroid-type antioxidant targeting the Keap1/Nrf2/ARE signaling pathway from the soft coral Dendronephthya gigantea. J. Nat. Prod. 2018, 81, 2567–2575. [Google Scholar] [CrossRef]
- Vanderah, D.J.; Djerassi, C. Marine natural products. Synthesis of four naturally occurring 20β-H cholanic acid derivatives. J. Org. Chem. 1978, 43, 1442–1448. [Google Scholar] [CrossRef]
- Matsumori, N.; Kaneno, D.; Murata, M.; Nakamura, H.; Tachibana, K. Stereochemical determination of acyclic structures based on carbon-proton spin-coupling constants. A method of configuration analysis for natural products. J. Org. Chem. 1999, 64, 866–876. [Google Scholar] [CrossRef]
- Ivanchina, N.V.; Kicha, A.A.; Kalinovsky, A.I.; Dmitrenok, P.S.; Dmitrenok, A.S.; Chaikina, E.L.; Stonik, V.A.; Gavagnin, M.; Cimino, G. Polar steroidal compounds from the far eastern starfish Henricia Leviuscula. J. Nat. Prod. 2006, 69, 224–228. [Google Scholar] [CrossRef]
- Sampson, N.; Neuwirt, H.; Puhr, M.; Klocker, H.; Eder, I.E. In vitro model systems to study androgen receptor signaling in prostate cancer. Endocr. Relat. Cancer 2013, 20, R49–R64. [Google Scholar] [CrossRef] [Green Version]
- Nelson, P.S. Targeting the androgen receptor in prostate cancer—A resilient foe. N. Engl. J. Med. 2014, 371, 1067–1069. [Google Scholar] [CrossRef] [Green Version]
- Iguchi, K.; Fujita, M.; Nagaoka, H.; Mitome, H.; Yamada, Y. Aragusterol Aa potent antitumor marine steroid from the Okinawan sponge of the genus Xestospongia. Tetrahedron Lett. 1993, 34, 6277–6280. [Google Scholar] [CrossRef]
- Koltun, V.M. Demospongian Sponges of the Northern and Far-Eastern Seas; Publisher of USSR Academy of Sciences: Moscow, Russia; Leningrad, Russia, 1959; pp. 1–236. (In Russian) [Google Scholar]
- Dyshlovoy, S.A.; Venz, S.; Hauschild, J.; Tabakmakher, K.M.; Otte, K.; Madanchi, R.; Walther, R.; Guzii, A.G.; Makarieva, T.N.; Shubina, L.K.; et al. Antimigrating activity of marine alkaloid monanchocidin A, proteome-based discovery and confirmation. Proteomics 2016, 16, 1590–1603. [Google Scholar] [CrossRef] [PubMed]
- Dyshlovoy, S.A.; Tabakmakher, K.M.; Hauschild, J.; Shchekaleva, R.K.; Otte, K.M.; Guzii, A.G.; Makarieva, T.N.; Kudryashova, E.K.; Shubina, L.K.; Bokemeyer, C.; et al. Anticancer activity of eight rare guanidine alkaloids isolated from marine sponge Monanchora pulchra. Mar. Drugs 2016, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Prinsep, M.R.; Blunt, J.W.; Munro, M.H.G. A new sterol sulfate from the marine sponge Stylopus australis. J. Nat. Prod. 1989, 52, 657–659. [Google Scholar] [CrossRef]
- Riccio, R.; Iorizzi, M.; Minale, L. Starfish saponins XXX. Isolation of sixteen steroidal glycosides and three polyhydroxysteroids from the mediterranean starfish Coscinasterias tenuispina. Bull. Soc. Chim. Belg. 1986, 95, 869–893. [Google Scholar] [CrossRef]





| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | δH (J in Hz) | |
| 1α | 1.39, m | 1.37, m | 1.37, m | 1.15, m | 1.37, m | 1.31, m | 1.15, m |
| 1β | 1.05, m | 2.10, m | 2.06, m | 1.90, m | 2.07, m | 2.12, dt (13.3, 3.3) | 1.89, dt (13.3, 3.3) |
| 2α | 1.82, m | 1.84, m | 1.82, m | 1.81, m | 1.83, m | 1.84, m | 1.82, m |
| 2β | 2.10, m | 2.12, m | 2.10, m | 2.11, m | 2.11, m | 2.21, m | 2.12, m |
| 3 | 4.27, ddd (11.5, 4.1, 3.0) | 4.29, ddd (11.5, 4.1, 3.0) | 4.27, ddd (11.5, 4.1, 3.0) | 4.18, ddd (12.2, 4.3, 3.3) | 4.28, ddd (11.5, 4.1, 3.0) | 4.19, ddd (12.0, 3.9, 3.1) | 4.18, ddd (11.7, 4.0, 3.1) |
| 4 | 3.55, br d (3.0) | 3.58, br d (3.0) | 3.56, br d (3.0) | 4.42, dd (3.3, 1.3) | 3.57, br d (3.0) | 4.40, dd (3.3, 1.3) | 4.42, dd (3.3, 1.3) |
| 5 | |||||||
| 6 | 3.17, br d (2.7) | 3.12, br d (2.7) | 3.17, br d (2.5) | 5.70, dd (5.0, 2.4) | 3.19, br d (2.5) | 5.59, dd (4.2, 3.0) | 5.70, dd (5.0, 2.4) |
| 7α 7β | 1.29, m 2.08, m | 1.35, m 2.19, m | 1.27, m 2.08, m | 1.68, ddd (18.2, 10.3, 2.3) 2.40, m | 1.32, m 2.39, m | 1.80, ddd (18.0, 9.8, 2.6) 2.53, ddd (18.8, 6.6, 4.3) | 1.68, ddd (18.0, 10.3, 2.3) 2.39, m |
| 8 | 1.43, m | 1.79, m | 1.42, m | 1.99, dd (10.8, 5.7) | 1.85, m | 2.42, m | 1.99, m |
| 9 | 0.68, dd (12.0, 4.5) | 0.78 dd, (11.5, 3.0) | 0.68, dd, (11.6, 4.7) | 0.98, m | 0.74, m | 1.01, m | 0.98, m |
| 11α 11 β | 1.40, m 1.43, m | 4.14, br q, (3.0) | 1.40, m 1.44, m | 1.50, m 1.52, m | 1.38, m 1.43, m | 4.29, br q, (3.4) | 1.49, m 1.51, m |
| 12α | 1.14, m | 1.30, m | 1.12, m | 1.18, m | 1.10, m | 1.36, m | 1.18, m |
| 12 β | 2.02, m | 2.23, dd, (13.3, 3.0) | 2.02, m | 2.03, dt (12.6, 3.6) | 1.97, dt (12.3, 3.7) | 2.22, m | 2.03, dt (12.5, 3.3) |
| 13 | |||||||
| 14 | 0.95, m | 0.94, m | 0.94, m | 0.90, m | 0.78, dd (11.3, 5.7) | 0.89, m | 0.90, m |
| 15 | 1.64, m 1.05, m | 1.65, m 1.15, m | 1.64, m 1.06, m | 4.15, ddd (7.9, 5.8, 2.2) | 4.15, ddd (8.1, 5.7, 2.3) | 4.18, ddd (7.8, 5.7, 2.2) | 4.15, ddd (7.7, 5.6, 2.0) |
| 16 | 1.86, m 1.36, m | 1.83, m 1.39, m | 1.86, m 1.35, m | 2.40, m 1.41, ddd (14.3, 10.4, 2.3) | 2.36, m 1.37, m | 2.37, m 1.41, m | 2.40, m 1.39, m |
| 17 | 1.08, m | 1.03, m | 1.08, m | 1.05, m | 1.02, m | 1.00, m | 1.07, m |
| 18 | 0.70, s | 0.93, s | 0.69, s | 1.00, s | 0.94, s | 1.20, s | 0.99, s |
| 19 | 1.16, s | 1.43, s | 1.16, s | 1.24, s | 1.19, s | 1.49, s | 1.24, s |
| 20 | 1.73, m | 1.72, m | 1.72, m | 1.88, m | 1.86, m | 1.88, m | 1.87, m |
| 21 | 0.94, d (6.7) | 0.96, d (6.7) | 0.95, d (6.7) | 1.02, d (6.7) | 0.99, d (6.7) | 1.04, d (6.7) | 0.97, d (6.7) |
| 22 | 1.41, m 1.04, m | 1.39, m 1.03, m | 1.48, m 0.98, m | 1.59, ddd (13.7, 10.3, 2.3) 1.11, m | 1.57, ddd (14.1, 10.5, 2.7) 1.10, m | 1.58, ddd (14.1, 10.5, 2.5) 1.11, m | 1.43, 1.07, m |
| 23 | 3.53, ddd (9.1, 7.3, 2.0) | 3.53, ddd (9.4, 7.3, 2.0) | 3.70, m | 4.13, br d (10.5) | 4.11, br d (10.5) | 4.13, br d (10.5) | 3.55, ddd (9.3, 7.1, 2.0) |
| 24 | 1.29, m | 1.28, m | 1.38, m 1.14, m | 1.31, m | |||
| 25 | 1.91, m | 1.91, m | 1.75, m | 2.26, septet (6,7) | 2.24, septet (6,7) | 2.25, septet (6,7) | 1.93, m |
| 26 | 0.82, d (6.6) | 0.82, d (6.6) | 0.90, d (6.8) | 1.06, d (6.9) | 1.05, d (6.8) | 1.06, d (6.8) | 0.83, d (6.9) |
| 27 | 0.91, d (6.6) | 0.91, d (6.6) | 0.91, d (6.8) | 1.08, d (6.9) | 1.07, d (6.8) | 1.08, d (6.8) | 0.92, d (6.9) |
| 28 | 0.74, d (6.8) | 0.74, d (6.8) | 5.03, t (1.2) 4.84, br s | 5.03, t (1.2) 4.84, br s | 5.03, t (1.2) 4.84, br s | 0.75, d |
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| δC, Type | δC, Type | δC, Type | δC, Type | δC, Type | δC, Type | δC, Type | |
| 1 | 39.2, CH2 | 40.2, CH2 | 39.2, CH2 | 39.2, CH2 | 39.2, CH2 | 38.2, CH2 | 39.4, CH2 |
| 2 | 23.7, CH2 | 24.1, CH2 | 23.7, CH2 | 24.6, CH2 | 23.7, CH2 | 24.0, CH2 | 24.4, CH2 |
| 3 | 80.1, CH | 79.9, CH | 80.1, CH | 82.5, CH | 80.1, CH | 82.0, CH | 82.1, CH |
| 4 | 77.7, CH | 78.0, CH | 77.7, CH | 77.6, CH | 77.7, CH | 76.9, CH | 77.6, CH |
| 5 | 66.4, C | 66.2, C | 66.4, C | 144.1, C | 66.3, C | 145.4, C | 144.1, C |
| 6 | 64.2, CH | 63.3, CH | 64.2, CH | 130.0, CH | 64.2, CH | 129.5, CH | 130.0, CH |
| 7 | 34.2, CH2 | 33.6, CH2 | 34.2, CH2 | 33.0, CH2 | 33.6, CH2 | 33.2, CH2 | 33.0, CH2 |
| 8 | 31.7, CH | 28.8, CH | 31.7, CH | 29.4, CH | 27.4, CH | 26.4, CH | 29.5, CH |
| 9 | 53.9, CH | 57.8, CH | 53.9, CH | 52.9, CH | 54.3, CH | 56.2, CH | 59.2, CH |
| 10 | 36.9, C | 37.9, C | 36.9, C | 38.0, C | 36.9, C | 38.6, C | 38.1, C |
| 11 | 23.1, CH2 | 69.5, CH | 23.1, CH2 | 22.1, CH | 23.1, CH2 | 69.5, CH | 22.2, CH |
| 12 | 41.8, CH2 | 51.1, CH2 | 41.8, CH2 | 43.0, CH2 | 43.1, CH2 | 52.3, CH2 | 43.0, CH2 |
| 13 | 44.1, C | 43.7, C | 44.1, C | 44.0, C | 43.9, C | 43.4, C | 44.0, C |
| 14 | 58.1, CH | 61.2, CH | 58.1, CH | 63.5, CH | 62.8, CH | 65.6, CH | 63.6, CH |
| 15 | 25.9, CH2 | 25.7, CH2 | 25.9, CH2 | 71.3, CH | 70.9, CH | 71.3, CH | 71.2, CH |
| 16 | 29.8, CH2 | 29.7, CH2 | 29.8, CH2 | 42.7, CH2 | 42.6, CH2 | 42.3, CH2 | 42.6, CH2 |
| 17 | 59.0, CH | 59.8, CH | 58.9, CH | 59.1, CH | 59.0, CH | 59.9, CH | 59.4, CH |
| 18 | 12.8, CH3 | 16.1, CH3 | 12.8, CH3 | 15.7, CH3 | 15.4, CH3 | 18.1, CH3 | 15.6, CH3 |
| 19 | 19.1, CH3 | 21.8, CH3 | 19.1, CH3 | 22.0, CH3 | 19.0, CH3 | 25.3, CH3 | 21.9, CH3 |
| 20 | 34.1, CH | 34.2, CH | 34.1, CH | 34.2, CH | 34.2, CH | 34.3, CH | 33.9, CH |
| 21 | 19.5, CH3 | 19.5, CH3 | 19.7, CH3 | 19.6, CH3 | 19.5, CH3 | 19.5, CH3 | 19.7, CH3 |
| 22 | 41.7, CH2 | 41.8, CH2 | 46.2, CH2 | 45.0, CH2 | 45.1, CH2 | 45.1, CH2 | 41.9, CH2 |
| 23 | 71.7, CH | 71.8, CH | 68.1, CH | 72.5, CH | 72.5, CH | 72.6, CH | 71.8, CH |
| 24 | 47.4, CH | 47.4, CH | 49.5, CH | 162.4, C | 162.3, C | 162.4, C | 47.4, CH |
| 25 | 29.6, CH | 29.6, CH | 26.4, CH | 32.2, CH | 32.1, CH | 32.2, CH | 29.6, CH |
| 26 | 22.5, CH3 | 22.5, CH3 | 24.4, CH3 | 24.4, CH3 | 24.4, CH3 | 24.4, CH3 | 22.5, CH3 |
| 27 | 18.3, CH3 | 18.4, CH3 | 23.3, CH3 | 23.6, CH3 | 23.7, CH3 | 23.7, CH3 | 18.4, CH3 |
| 28 | 11.3, CH3 | 11.3, CH3 | 106.8, CH2 | 106.8, CH2 | 106.8, CH2 | 11.4, CH3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shubina, L.K.; Makarieva, T.N.; Denisenko, V.A.; Popov, R.S.; Dyshlovoy, S.A.; Grebnev, B.B.; Dmitrenok, P.S.; von Amsberg, G.; Stonik, V.A. Gracilosulfates A–G, Monosulfated Polyoxygenated Steroids from the Marine Sponge Haliclona gracilis. Mar. Drugs 2020, 18, 454. https://doi.org/10.3390/md18090454
Shubina LK, Makarieva TN, Denisenko VA, Popov RS, Dyshlovoy SA, Grebnev BB, Dmitrenok PS, von Amsberg G, Stonik VA. Gracilosulfates A–G, Monosulfated Polyoxygenated Steroids from the Marine Sponge Haliclona gracilis. Marine Drugs. 2020; 18(9):454. https://doi.org/10.3390/md18090454
Chicago/Turabian StyleShubina, Larisa K., Tatyana N. Makarieva, Vladimir A. Denisenko, Roman S. Popov, Sergey A. Dyshlovoy, Boris B. Grebnev, Pavel S. Dmitrenok, Gunhild von Amsberg, and Valentin A. Stonik. 2020. "Gracilosulfates A–G, Monosulfated Polyoxygenated Steroids from the Marine Sponge Haliclona gracilis" Marine Drugs 18, no. 9: 454. https://doi.org/10.3390/md18090454
APA StyleShubina, L. K., Makarieva, T. N., Denisenko, V. A., Popov, R. S., Dyshlovoy, S. A., Grebnev, B. B., Dmitrenok, P. S., von Amsberg, G., & Stonik, V. A. (2020). Gracilosulfates A–G, Monosulfated Polyoxygenated Steroids from the Marine Sponge Haliclona gracilis. Marine Drugs, 18(9), 454. https://doi.org/10.3390/md18090454