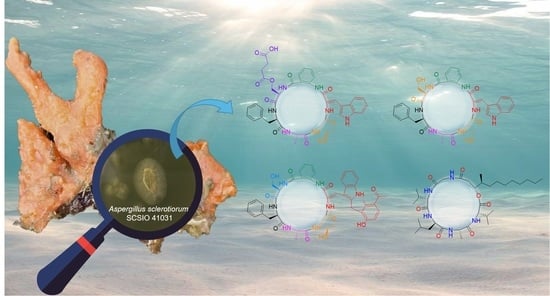
Cyclic Peptides from the Soft Coral-Derived Fungus Aspergillus sclerotiorum SCSIO 41031
Abstract
:1. Introduction
2. Results
2.1. Structure Elucidate
2.2. Bioassays
2.3. Molecular Docking
3. Materials and Methods General Experimental Procedures
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation and Extraction
3.4. Isolation and Purification
3.5. Marfey’s Analysis of 2 and 3
3.6. X-ray Crystallographic Analysis
3.7. Bioactivity Assay
3.8. Molecular Docking Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine Natural Products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef] [PubMed]
- Darcel, L.; Das, S.; Bonnard, I.; Banaigs, B.; Inguimbert, N. Thirtieth Anniversary of the Discovery of Laxaphycins. Intriguing Peptides Keeping a Part of Their Mystery. Mar. Drugs 2021, 19, 473. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tao, H.; Lei, X.; Zhang, H.; Zhou, X.; Liu, Y.; Li, Y.; Yang, B. Arthriniumsteroids A-D, Four New Steroids from the Soft Coral-Derived Fungus Simplicillium lanosoniveum SCSIO41212. Steroids 2021, 171, 108831. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Liu, D.; Shao, Z.; Huang, J.; Fan, A.; Lin, W. Chlorinated Metabolites with Antibacterial Activities from a Deep-Sea-Derived Spiromastix Fungus. RSC Adv. 2021, 11, 29661–29667. [Google Scholar] [CrossRef]
- Zhang, H.; Lei, X.-X.; Shao, S.; Zhou, X.; Li, Y.; Yang, B. Azaphilones and Meroterpenoids from the Soft Coral-Derived Fungus Penicillium glabrum Glmu003. Chem. Biodivers. 2021, 18, e2100663. [Google Scholar] [CrossRef] [PubMed]
- Dayanidhi, D.L.; Thomas, B.C.; Osterberg, J.S.; Vuong, M.; Vargas, G.; Kwartler, S.K.; Schmaltz, E.; Dunphy-Daly, M.M.; Schultz, T.F.; Rittschof, D.; et al. Exploring the Diversity of the Marine Environment for New Anti-Cancer Compounds. Front. Mar. Sci. 2021, 7, 4766. [Google Scholar] [CrossRef]
- Esposito, R.; Ruocco, N.; Viel, T.; Federico, S.; Zupo, V.; Costantini, M. Sponges and Their Symbionts as a Source of Valuable Compounds in Cosmeceutical Field. Mar. Drugs 2021, 19, 444. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Williams, P.G.; Kwon, H.C.; Jensen, P.R.; Fenical, W. Lucentamycins A−D, Cytotoxic Peptides from the Marine-Derived Actinomycete Nocardiopsis lucentensis. J. Nat. Prod. 2007, 70, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Huang, H.; Chen, Y.; Tan, J.; Song, Y.; Zou, J.; Tian, X.; Hua, Y.; Ju, J.; Marthiapeptide, A. An Anti-Infective and Cytotoxic Polythiazole Cyclopeptide from a 60 L Scale Fermentation of the Deep Sea-Derived Marinactinospora Thermotolerans SCSIO 00652. J. Nat. Prod. 2012, 75, 2251–2255. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Xu, Z.; Wang, Y.; Hong, K.; Liu, P.; Zhu, W. Cyclic Tripeptides from the Halotolerant Fungus Aspergillus sclerotiorum PT06–1. J. Nat. Prod. 2010, 73, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, K.; Inaba, S.; Takagi, M.; Shin-Ya, K. JBIR-15, a New Aspochracin Derivative, Isolated from a Sponge-Derived Fungus, Aspergillus sclerotiorum Huber Sp080903f04. Biosci. Biotechnol. Biochem. 2009, 73, 1898–1900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, F.-H.; Liang, X.; Cheng, X.; Ling, J.; Dong, J.-D.; Qi, S.-H. Antifungal peptides from the marine gorgonian-associated fungus Aspergillus sp. SCSIO41501. Phytochemistry 2021, 192, 112967. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhu, H.; Hong, K.; Wang, Y.; Liu, P.; Wang, X.; Peng, X.; Zhu, W. Novel cyclic hexapeptides from marine-derived fungus, Aspergillus sclerotiorum PT06–1. Org. Lett. 2009, 11, 5262–5265. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Lin, Y.; Pang, X.; Luo, X.; Salendra, L.; Wang, J.; Zhou, X.; Lu, Y.; Yang, B.; Liu, Y. Peptides from the soft coral-associated fungus Simplicillium sp. SCSIO41209. Phytochemistry 2018, 154, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Elbanna, A.H.; Khalil, Z.G.; Bernhardt, P.V.; Capon, R.J. Scopularides revisited: Molecular networking guided exploration of lipodepsipeptides in Australian marine fish gastrointestinal tract-derived fungi. Mar. Drugs 2019, 17, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rydberg, E.H.; Brumshtein, B.; Greenblatt, H.M.; Wong, D.M.; Shaya, D.; Williams, L.D.; Carlier, P.R.; Pang, Y.-P.; Silman, I.; Sussman, J.L. Complexes of alkylene-linked tacrine dimers with torpedo californica acetylcholinesterase: Binding of bis5-tacrine produces a dramatic rearrangement in the active-site gorge. J. Med. Chem. 2006, 49, 5491–5500. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L.; Courtney, K.D.; Andres, V.; Feather-Stone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Soliseptide, A. A cyclic hexapeptide possessing piperazic acid groups from Streptomyces solisilvae HNM30702. Org. Lett. 2018, 20, 1371–1374. [Google Scholar]
- Sang, B.H.; Shin, Y.J.; Hyon, J.Y.; Wee, W.R. Cytotoxicity of Voriconazole on Cultured Human Corneal Endothelial Cells. Antimicrob. Agents. Chemother. 2011, 55, 4519–4523. [Google Scholar]
- Dai, Y.; Li, K.; She, J.; Zeng, Y.; Wang, H.; Liao, S.; Lin, X.; Yang, B.; Wang, J.; Tao, H.; et al. Lipopeptide Epimers and a Phthalide Glycerol Ether with AChE Inhibitory Activities from the Marine-Derived Fungus Cochliobolus lunatus SCSIO41401. Mar. Drugs 2020, 18, 547. [Google Scholar] [CrossRef] [PubMed]






| Position | Sclerotide C (1) | Position | Sclerotide D (2) | ||||
|---|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | ||||
| Thr | 1 | 170.3 | Ser | 1 | 169.7 | ||
| 2 | 59.0 | 4.32, m | 2 | 56.8 | 4.55, td (7.2, 4.2) | ||
| 3 | 65.5 | 4.44, m | 3 | 60.7 | 4.04, dd (11.2, 8.7) | ||
| 4 | 20.8 | 1.17, d (6.4) | 3.96, dd (11.3, 4.3) | ||||
| NH | 7.33, d (8.4) | NH | 7.13, d (8.1) | ||||
| Ala | 5 | 172.6 | Ala | 4 | 172.5 | ||
| 6 | 49.6 | 4.22, m | 5 | 49.2 | 4.25, m | ||
| 7 | 16.7 | 1.28, d (7.3) | 6 | 16.3 | 1.25, d (7.3) | ||
| NH | 8.74, d (6.1) | NH | 8.62, d (6.5) | ||||
| Phe | 8 | 171.5 | Phe | 7 | 171.8 | ||
| 9 | 54.4 | 4.63, “q” like (7.6) | 8 | 54.5 | 4.53, “q” like (7.6) | ||
| 10 | 35.6 | 2.89, dd (14.1, 7.9) | 9 | 35.1 | 2.93, dd (14.0, 7.3) | ||
| 2.93, dd (14.1, 7.1) | 2.88, dd (14.0, 7.9) | ||||||
| 11 | 137.6 | 10 | 137.5 | ||||
| 12 | 129.0 | 7.20, d (7.4) | 11 | 129.0 | 7.20, d (7.2) | ||
| 13 | 128.1 | 7.24, t (7.4) | 12 | 128.1 | 7.23, m | ||
| 14 | 126.3 | 7.18, t (7.4) | 13 | 126.2 | 7.17, m | ||
| 15 | 128.1 | 7.24, t (7.4) | 14 | 128.1 | 7.23, m | ||
| 16 | 129.0 | 7.20, d (7.4) | 15 | 129.0 | 7.20, d (7.2) | ||
| NH | 8.43, d (7.5) | NH | 8.26, d (7.0) | ||||
| BASE | 17 | 168.8 | Ser | 16 | 170.8 | ||
| 18 | 54.4 | 4.51, td (7.2, 4.2) | 17 | 59.2 | 4.18, q (5.8) | ||
| 19 | 63.0 | 4.26, dd, (11.3, 7.9) | 18 | 60.8 | 3.70, d (5.7) | ||
| 4.31, m | |||||||
| 39 | 172.1 | NH | 8.65, d (5.5) | ||||
| 40 | 28.6 | 2.45, ovl a | |||||
| 41 | 28.6 | 2.45, ovl a | |||||
| 42 | 173.4 | ||||||
| NH | 8.98, d (6.2) | ||||||
| AA | 20 | 169.3 | AA | 19 | 169.5 | ||
| 21 | 122.0 | 20 | 119.6 | ||||
| 22 | 129.2 | 7.79, dd (7.8, 1.1) | 21 | 129.1 | 7.96, dd (8.1, 1.1) | ||
| 23 | 122.7 | 7.23, m | 22 | 122.2 | 7.20, m | ||
| 24 | 132.2 | 7.59, ddd (8.2, 7.8, 1.0) | 23 | 132.6 | 7.59, ddd (8.4, 6.6, 1.0) | ||
| 25 | 121.0 | 8.58, d (8.2) | 24 | 119.6 | 8.79, d (8.3) | ||
| 26 | 138.6 | 25 | 139.7 | ||||
| NH | 10.89, s | NH | 11.75, s | ||||
| ∆-Trp | 27 | 163.5 | ∆-Trp | 26 | 163.4 | ||
| 28 | 122.2 | 27 | 121.8 | ||||
| 29 | 125.9 | 7.96, s | 28 | 126.3 | 7.97, s | ||
| 30 | 108.6 | 29 | 108.7 | ||||
| 31 | 127.4 | 30 | 127.4 | ||||
| 32 | 117.7 | 7.76, d (7.8) | 31 | 117.7 | 7.76, d (7.7) | ||
| 33 | 120.4 | 7.16, m | 32 | 120.4 | 7.16, m | ||
| 34 | 122.0 | 7.18, m | 33 | 122.2 | 7.20, m | ||
| 35 | 112.0 | 7.42,d (7.9) | 34 | 111.9 | 7.42, d (7.9) | ||
| 36 | 135.5 | 35 | 135.5 | ||||
| 37-NH | 11.95, d (1.9) | 36-NH | 11.97, d (2.0) | ||||
| 38 | 128.7 | 8,00 d (2.8) | 37 | 128.7 | 8.03, d (2.8) | ||
| NH | 8.79, s | NH | 8.82, s | ||||
| Position | Sclerotides E (3) | Position | Scopularide I (4) | ||||
|---|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | ||||
| Thr | 1 | 169.6 | Val | 1 | 170.8 | ||
| 2 | 57.5 | 4.35, dd (9.4, 3.2) | 2 | 57.7 | 4.09, ovl a | ||
| 3 | 66.2 | 4.17, td (6.5, 3.7) | 3 | 29.6 | 2.07, m | ||
| 4 | 19.9 | 1.01, d (6.4) | 4 | 18.8 | 0.87, ovl a | ||
| 3-OH | 4.80, br s | 5 | 17.4 | 0.86, ovl a | |||
| NH | 7.32, d (8.6) | NH | 7.42, br s | ||||
| Ala | 5 | 171.9 | Ala | 6 | 171.9 | ||
| 6 | 48.1 | 4.47, m | 7 | 48.0 | 4.19, m | ||
| 7 | 16.6 | 1.14, d (7.2) | 8 | 17.5 | 1.22, d (7.1) | ||
| NH | 8.39, d (8.5) | NH | 8.00, d (7.8) | ||||
| Phe | 8 | 170.3 | Leu | 9 | 171.4 | ||
| 9 | 54.4 | 4.55, “q” like | 10 | 51.9 | 4.04, ovl a | ||
| 10 | 35.6 | 2.90, dd (13.7, 7.8) | 11 | 38.7 | 1.49, m | ||
| 3.02, dd (13.8, 7.5) | 12 | 24.2 | 1.64, m | ||||
| 11 | 137.6 | 13 | 23.0 | 0.89, ovl a | |||
| 12 | 129.1 | 7.27, d (7,3) | 14 | 21.0 | 0.81, ovl a | ||
| 13 | 128.1 | 7.25, m | NH | 8.65, d (6.6) | |||
| 14 | 126.3 | 7.2, m | Val | 15 | 171.7 | ||
| 15 | 128.1 | 7.25, m | 16 | 58.8 | 4.05, ovl a | ||
| 16 | 129.1 | 7.27, d (7,3) | 17 | 29.5 | 1.85, m | ||
| NH | 8.31, d (7.8) | 18 | 19.0 | 0.88, ovl a | |||
| Ser | 17 | 170.8 | 19 | 19.1 | 0.88, ovl a | ||
| 18 | 57.8 | 4.23, q (6.8) | NH | 8.07, d (7.6) | |||
| 19 | 61.1 | 3.67, m | Gly | 20 | 169.1 | ||
| 19-OH | 5.15, m | 21 | 42.0 | 4.08, ovl a | |||
| NH | 8.31, d (7.8) | 3.45, dd (17.1, 3.7) | |||||
| AA | 20 | 168.1 | NH | 7.90, dd (6.6, 3.9) | |||
| 21 | 123.1 | HMLA | 22 | 169.9 | |||
| 22 | 129.7 | 7.94, d (7.9) | 23 | 37.5 | 2.53, dd (15.6, 9.6) | ||
| 23 | 129.2 | 7.27, m | 2.24, m | ||||
| 24 | 132.1 | 7.58, ddd (8.4, 6.6, 1.0) | 24 | 75.5 | 4.91, ddd (9.4, 3.8, 2.3) | ||
| 25 | 122.2 | 8.36, m | 25 | 36.3 | 1.67, m | ||
| 26 | 138.3 | 26 | 31.6 | 1.36, m | |||
| NH | 10.86, br s | 1.02, m | |||||
| ADPAT | 27 | 164.2 | 27 | 26.6 | 1.28, ovl a | ||
| 28 | 123.2 | 1.18, ovl a | |||||
| 29 | 126.2 | 7.87, s | 28 | 28.7 | 1.23, ovl a | ||
| 30 | 106.8 | 29 | 28.9 | 1.23, ovl a | |||
| 31 | 125.3 | 30 | 29.3 | 1.23, ovl a | |||
| 32 | 120.7 | 7.34, m | 31 | 31.3 | 1.23, ovl a | ||
| 33 | 120.9 | 7.02, m | 32 | 22.1 | 1.27, ovl a | ||
| 34 | 120.1 | 7.02, m | 33 | 14.0 | 0.85, s | ||
| 35 | 112.6 | 7.34, m | 34 | 14.8 | 0.83, ovl a | ||
| 36 | 135.9 | ||||||
| 37-NH | 10.78, s | ||||||
| 38 | 141.6 | ||||||
| NH | 9.32, s | ||||||
| 39 | 19.8 | 4.05, d (14.7) | |||||
| 40 | 110.9 | ||||||
| 41 | 162.4 | ||||||
| 41-OH | 13.27, s | ||||||
| 42 | 111.5 | ||||||
| 43 | 131.9 | 7.75, d (8.9) | |||||
| 44 | 107.5 | 6.53, d (8.8) | |||||
| 45 | 162.8 | ||||||
| 45-OH | 11.01, s | ||||||
| 46 | 203.3 | ||||||
| 47 | 26.1 | 2.54, s | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, J.; Chen, Y.; Chen, W.; Wang, J.; Zhou, X.; Yang, B.; Liu, Y. Cyclic Peptides from the Soft Coral-Derived Fungus Aspergillus sclerotiorum SCSIO 41031. Mar. Drugs 2021, 19, 701. https://doi.org/10.3390/md19120701
Long J, Chen Y, Chen W, Wang J, Zhou X, Yang B, Liu Y. Cyclic Peptides from the Soft Coral-Derived Fungus Aspergillus sclerotiorum SCSIO 41031. Marine Drugs. 2021; 19(12):701. https://doi.org/10.3390/md19120701
Chicago/Turabian StyleLong, Jieyi, Yaqi Chen, Weihao Chen, Junfeng Wang, Xuefeng Zhou, Bin Yang, and Yonghong Liu. 2021. "Cyclic Peptides from the Soft Coral-Derived Fungus Aspergillus sclerotiorum SCSIO 41031" Marine Drugs 19, no. 12: 701. https://doi.org/10.3390/md19120701
APA StyleLong, J., Chen, Y., Chen, W., Wang, J., Zhou, X., Yang, B., & Liu, Y. (2021). Cyclic Peptides from the Soft Coral-Derived Fungus Aspergillus sclerotiorum SCSIO 41031. Marine Drugs, 19(12), 701. https://doi.org/10.3390/md19120701