A Crustin from Hydrothermal Vent Shrimp: Antimicrobial Activity and Mechanism
Abstract
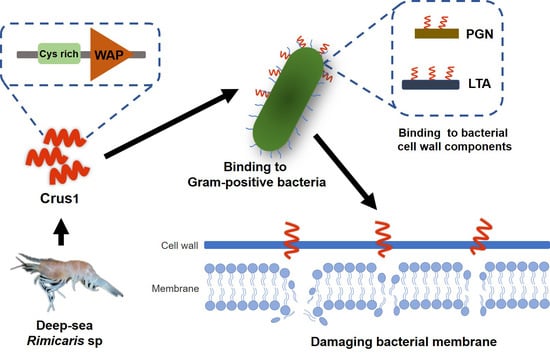
:1. Introduction
2. Results
2.1. Sequence and Structure Characterization of Crus1
2.2. Antimicrobial Activity of rCrus1 and Its Dependence on Temperature, pH, and Disulfide Bonds
2.3. Binding of rCrus1 to Bacterial Cell Wall Components and Its Effect on Bactericidal Activity
2.4. Effects of rCrus1 on the Morphology and Membrane Integrity of Bacteria
2.5. The Conserved Cysteine Residues in the WAP Domain Are Essential to the Antimicrobial Activity of rCrus1
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Bioinformatics Analysis and Structural Modeling of Crus1
4.3. Protein Expression and Purification
4.4. Antibacterial Activity Assay
4.5. Protein Binding to Bacteria and Cell Wall Components
4.6. Electron Microscopy and PI Staining Assay
4.7. Bacterial Cytoplasmic Membrane Depolarization
4.8. Protoplast Preparation and Lysis Assay
4.9. Immunofluorescence Microscopy
4.10. Circular Dichroism (CD) Spectroscopy
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, Y.; Ren, Q. Research progress in innate immunity of freshwater crustaceans. Dev. Comp. Immunol. 2019, 104, 103569. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [Green Version]
- Brown, K.L.; Hancock, R.E. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 2006, 18, 24–30. [Google Scholar] [CrossRef]
- Benincasa, M.; Runti, G.; Mardirossian, M.; Scocchi, M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top. Med. Chem. 2016, 16, 76–88. [Google Scholar]
- Giuliani, A.; Pirri, G.; Nicoletto, S.F. Antimicrobial peptides: An overview of a promising class of therapeutics. Cent. Eur. J. Biol. 2007, 2, 1–33. [Google Scholar] [CrossRef]
- Hancock, R.E.; Hancock, R.E.W. Cationic peptides: Effectors in innate immunity & novel antimicrobials. Lancet Infect. Dis. 2001, 1, 156–164. [Google Scholar] [PubMed]
- Boman, H.G. Antibacterial peptides: Basic facts and emerging concepts. J. Intern. Med. 2010, 254, 197–215. [Google Scholar] [CrossRef] [PubMed]
- Essig, A.; Hofmann, D.; Munch, D.; Gayathri, S.; Kunzler, M.; Kallio, P.T.; Sahl, H.G.; Wider, G.; Schneider, T.; Aebi, M. Copsin, a Novel Peptide-based Fungal Antibiotic Interfering with the Peptidoglycan Synthesis. J. Biol. Chem. 2014, 289, 34953–34964. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Rozek, A.; Hancock, R.E.W. Interaction of Cationic Antimicrobial Peptides with Model Membranes. J. Biol. Chem. 2001, 276, 35714–35722. [Google Scholar] [CrossRef] [Green Version]
- Arias, M.; Jensen, K.V.; Nguyen, L.T.; Storey, D.G.; Vogel, H.J. Hydroxy-tryptophan containing derivatives of tritrpticin: Modification of antimicrobial activity and membrane interactions. Biochim. Biophys. Acta Biomembr. 2015, 1848, 277–288. [Google Scholar] [CrossRef] [Green Version]
- Novkovic, M. Selective antimicrobial activity and mode of action of adepantins, glycine-rich peptide antibiotics based on anuran antimicrobial peptide sequences. Biochim. Biophys. Acta 2013, 1828, 1004–1012. [Google Scholar]
- Hilde, U.; Ørjan, S.; Haukland, H.H.; Manuela, K.; Vorland, L.H. Lactoferricin B inhibits bacterial macromolecular synthesis in Escherichia coli and Bacillus subtilis. FEMS Microbiol. Lett. 2004, 237, 377–384. [Google Scholar]
- Haney, E.F.; Petersen, A.P.; Lau, C.K.; Jing, W.; Storey, D.G.; Vogel, H.J. Mechanism of action of puroindoline derived tryptophan-rich antimicrobial peptides. Biochim. Biophys. Acta 2013, 1828, 1802–1813. [Google Scholar] [CrossRef] [Green Version]
- Schneider, T.; Kruse, T.; Wimmer, R.; Wiedemann, I.; Kristensen, H.-H. Plectasin, a Fungal Defensin, Targets the Bacterial Cell Wall Precursor Lipid II. Science 2010, 328, 1168–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, V.J.; Fernandes, J.M.O.; Kemp, G.D.; Hauton, C. Crustins: Enigmatic WAP domain-containing antibacterial proteins from crustaceans. Dev. Comp. Immunol. 2008, 32, 758–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauton, C.; Brockton, V.; Smith, V.J. Cloning of a crustin-like, single whey-acidic-domain, antibacterial peptide from the haemocytes of the European lobster, Homarus gammarus, and its response to infection with bacteria. Mol. Immunol. 2006, 43, 1490–1496. [Google Scholar] [CrossRef]
- Sallenave, J.-M. The role of secretory leukocyte proteinase inhibitor and elafin (elastase-specific inhibitor/skin-derived antileukoprotease) as alarm antiproteinases in inflammatory lung disease. Respir. Res. 2000, 1, 5. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, J.X.; Wang, Y.; Fang, W.H.; Wang, Y.; Zhou, J.F.; Zhao, S.; Li, X.C. Newly identified type II crustin (SpCrus2) in Scylla paramamosain contains a distinct cysteine distribution pattern exhibiting broad antimicrobial activity. Dev. Comp. Immunol. 2018, 84, 1–13. [Google Scholar] [CrossRef]
- Supungul, P.; Tang, S.; Maneeruttanarungroj, C.; Rimphanitchayakit, V.; Hirono, I.; Aoki, T.; Tassanakajon, A. Cloning, expression and antimicrobial activity of crustinPm1, a major isoform of crustin, from the black tiger shrimp Penaeus monodon. Dev. Comp. Immunol. 2008, 32, 61–70. [Google Scholar] [CrossRef]
- Arockiaraj, J.; Gnanam, A.J.; Muthukrishnan, D.; Gudimella, R.; Milton, J.; Singh, A.; Muthupandian, S.; Kasi, M.; Bhassu, S. Crustin, a WAP domain containing antimicrobial peptide from freshwater prawn Macrobrachium rosenbergii: Immune characterization. Fish Shellfish Immunol. 2013, 34, 109–118. [Google Scholar] [CrossRef]
- Jobstvogt, N.; Hanley, N.; Hynes, S.; Kenter, J.; Witte, U. Twenty thousand sterling under the sea: Estimating the value of protecting deep-sea biodiversity. Ecol. Econ. 2014, 97, 10–19. [Google Scholar] [CrossRef] [Green Version]
- Folkersen, M.V.; Fleming, C.M.; Hasan, S. The economic value of the deep sea: A systematic review and meta-analysis. Mar. Policy 2018, 94, 71–80. [Google Scholar] [CrossRef]
- Rodrigo, A.P.; Costa, P.M. The hidden biotechnological potential of marine invertebrates: The Polychaeta case study. Environ. Res. 2019, 173, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Costantini, M.; Sansone, C.; Lauritano, C.; Ruocco, N.; Ianora, A. Marine microorganisms as a promising and sustainable source of bioactive molecules. Mar. Environ. Res. 2016, 128, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Iván, H.Á.; Marie-Anne, C.B.; Florence, P.; Sébastien, D. Morphology of First Zoeal Stage of Four Genera of Alvinocaridid Shrimps from Hydrothermal Vents and Cold Seeps: Implications for Ecology, Larval Biology and Phylogeny. PLoS ONE 2015, 10, e0144657. [Google Scholar]
- Zhang, J.; Sun, Q.L.; Luan, Z.D.; Lian, C.; Sun, L. Comparative transcriptome analysis of Rimicaris sp. reveals novel molecular features associated with survival in deep-sea hydrothermal vent. Sci. Rep. 2017, 7, 2000. [Google Scholar] [CrossRef] [PubMed]
- Bloa, S.L.; Boidin-Wichlacz, C.; Cueff-Gauchard, V.; Rosa, R.D.; Tasiemski, A. Antimicrobial Peptides and Ectosymbiotic Relationships: Involvement of a Novel Type IIa Crustin in the Life Cycle of a Deep-Sea Vent Shrimp. Front. Immunol. 2020, 11, 1511. [Google Scholar] [CrossRef]
- Torrent, M.; Navarro, S.; Moussaoui, M.; Nogués, M.V.; Boix, E. Eosinophil Cationic Protein High-Affinity Binding to Bacteria-Wall Lipopolysaccharides and Peptidoglycans. Biochemistry 2008, 47, 3544–3555. [Google Scholar] [CrossRef]
- Bellemare, A.; Vernoux, N.; Morin, S.; Gagné, S.M.; Bourbonnais, Y. Structural and antimicrobial properties of human pre-elafin/trappin-2 and derived peptides against Pseudomonas aeruginosa. BMC Microbiol. 2010, 10, 253. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.F.; Wang, J.X. The antimicrobial peptides of the immune response of shrimp. Invertebr. Surviv. J. 2008, 5, 4. [Google Scholar]
- Imjongjirak, C.; Amparyup, P.; Tassanakajon, A.; Sittipraneed, S. Molecular cloning and characterization of crustin from mud crab Scylla paramamosain. Mol. Biol. Rep. 2009, 36, 841–850. [Google Scholar] [CrossRef]
- Bandeira, P.T.; Vernal, J.; Matos, G.M.; Farias, N.D.; Rosa, R.D. A Type IIa crustin from the pink shrimp Farfantepenaeus paulensis (crusFpau) is constitutively synthesized and stored by specific granule-containing hemocyte subpopulations. Fish Shellfish Immunol. 2019, 97, 294–299. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, R.R.; Fan, Z.X.; Zhao, X.F.; Wang, X.W. Characterization of a type-I crustin with broad-spectrum antimicrobial activity from red swamp crayfish Procambarus clarkii. Dev. Arative Immunol. 2016, 61, 145–153. [Google Scholar] [CrossRef]
- Krusong, K.; Poolpipat, P.; Supungul, P.; Tassanakajon, A. A comparative study of antimicrobial properties of crustinPm1 and crustinPm7 from the black tiger shrimp Penaeus monodon. Dev. Comp. Immunol. 2011, 36, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Dang, W.; Yan, J.; Chen, R.; Liu, X.; Yan, W.; Zhang, B.; Xie, J.; Zhang, J.; Wang, R. Membrane Perturbation Action Mode and Structure-Activity Relationships of Protonectin, a Novel Antimicrobial Peptide from the Venom of the Neotropical Social Wasp. Chem. Funct. Proteins 2013, 57, 4632–4639. [Google Scholar] [CrossRef] [Green Version]
- Koprivnjak, T.; Weidenmaier, C.; Peschel, A.; Weiss, J.P. Wall Teichoic Acid Deficiency in Staphylococcus aureus Confers Selective Resistance to Mammalian Group IIA Phospholipase A2 and Human β-Defensin 3. Infect. Immun. 2008, 76, 2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nermina, M.; Karl, L. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar]
- Brown, S.; John Maria, S.P.; Suzanne, W. Wall Teichoic Acids of Gram-Positive Bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef] [Green Version]
- Oppedijk, S.F.; Martin, N.I.; Breukink, E. Hit’em where it hurts: The growing and structurally diverse family of peptides that target lipid-II. Biochim. Biophys. Acta 2016, 1858, 947–957. [Google Scholar] [CrossRef]
- Kruijff, B.D.; Dam, V.V.; Breukink, E. Lipid II: A central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot. Essent. Fat. Acids 2008, 79, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.I.; Breukink, E. Expanding role of lipid II as a target for lantibiotics. Future Microbiol. 2007, 2, 513–525. [Google Scholar] [CrossRef]
- Gu, H.J.; Sun, Q.L.; Jiang, S.; Zhang, J.; Sun, L. First characterization of an anti-lipopolysaccharide factor (ALF) from hydrothermal vent shrimp: Insights into the immune function of deep-sea crustacean ALF. Dev. Comp. Immunol. 2018, 84, 382–395. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, C.; Gu, H.J.; Zhang, J.; Sun, L. Characterization of the Genome Feature and Toxic Capacity of a Bacillus wiedmannii Isolate From the Hydrothermal Field in Okinawa Trough. Front. Cell. Infect. Microbiol. 2019, 9, 370. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.J.; Miao, Y.Q.; Hu, L.P.; Kai, W.; Zhu, R. Immunization of mice against alpha, beta, and epsilon toxins of Clostridium perfringens using recombinant rCpa-b-x expressed by Bacillus subtilis. Mol. Immunol. 2020, 123, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.C.; Long, H.; Zhang, J.; Zhao, Y.; Sun, L. Characterization of a deepsea Bacillus toyonensisisolate: Genomicand pathogenicfeatures. Front. Cell. Infect. Microbiol. 2021, 11, 107. [Google Scholar] [CrossRef]
- Nielsen, H.; Tsirigos, K.D.; Brunak, S.; von Heijne, G. A brief history of protein sorting prediction. Protein J. 2019, 38, 200–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.; Zhang, P.; Zhang, T.; Sun, L. IL-34 regulates the inflammatory response and anti-bacterial immune defense of Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2020, 104, 228–236. [Google Scholar] [CrossRef]
- Zhang, B.C.; Sun, L. Tongue sole (Cynoglossus semilaevis) prothymosin alpha: Cytokine-like activities associated with the intact protein and the C-terminal region that lead to antiviral immunity via Myd88-dependent and -independent pathways respectively. Dev. Comp. Immunol. 2015, 53, 96–104. [Google Scholar] [CrossRef]
- Rončević, T.; Čikeš-Čulić, V.; Maravić, A.; Capanni, F.; Gerdol, M.; Pacor, S.; Tossi, A.; Giulianini, P.; Pallavicini, A.; Manfrin, C. Identification and functional characterization of the astacidin family of proline-rich host defence peptides (PcAst) from the red swamp crayfish (Procambarus clarkii, Girard 1852). Dev. Comp. Immunol. 2020, 105, 103574. [Google Scholar] [CrossRef]
- Li, W.R.; Guan, X.L.; Jiang, S.; Sun, L. The novel fish miRNA pol-miR-novel_171 and its target gene FAM49B play a critical role in apoptosis and bacterial infection. Dev. Comp. Immunol. 2020, 106, 103616. [Google Scholar] [CrossRef] [PubMed]
- Amparyup, P.; Sutthangkul, J.; Charoensapsri, W.; Tassanakajon, A. Pattern recognition protein binds to lipopolysaccharide and β-1,3-glucan and activates shrimp prophenoloxidase system. J. Biol. Chem. 2016, 291, 10949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Jiang, S.; Sun, L. A Fish Galectin-8 Possesses Direct Bactericidal Activity. Int. J. Mol. Sci. 2020, 22, 376. [Google Scholar] [CrossRef] [PubMed]
- Li, M.F.; Jia, B.B.; Sun, Y.Y.; Sun, L. The Translocation and Assembly Module (TAM) of Edwardsiella tarda Is Essential for Stress Resistance and Host Infection. Front. Microbiol. 2020, 11, 1743. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Gu, H.; Zhao, Y.; Sun, L. Teleost Gasdermin E Is Cleaved by Caspase 1, 3, and 7 and Induces Pyroptosis. J. Immunol. 2019, 203, ji1900383. [Google Scholar] [CrossRef] [PubMed]






| Bacteria | MIC (μM) | MBC (μM) |
|---|---|---|
| Gram-positive | ||
| Bacillus subtilis WB800N | 20 | 40 |
| Bacillus subtilis G7 | 40 | 40 |
| Bacillus wiedmannii SR52 | 20 | 40 |
| Bacillus cereus MB1 | 20 | 40 |
| Bacillus toyonensis P18 | 30 | 60 |
| Bacillus sp | 30 | 60 |
| Micrococcus luteus | 2.5 | 5 |
| Staphylococcus aureus | 10 | 20 |
| Streptococcus iniae | 15 | 30 |
| Gram-negative | ||
| Escherichia coli | — | — |
| Vibrio harveyi | — | — |
| Edwardsiella tarda | — | — |
| Vibrio anguillarum | — | — |
| Pseudoalteromonas sp | ≥200 | — |
| Pseudomonas fluorescens | — | — |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhang, J.; Sun, Y.; Sun, L. A Crustin from Hydrothermal Vent Shrimp: Antimicrobial Activity and Mechanism. Mar. Drugs 2021, 19, 176. https://doi.org/10.3390/md19030176
Wang Y, Zhang J, Sun Y, Sun L. A Crustin from Hydrothermal Vent Shrimp: Antimicrobial Activity and Mechanism. Marine Drugs. 2021; 19(3):176. https://doi.org/10.3390/md19030176
Chicago/Turabian StyleWang, Yujian, Jian Zhang, Yuanyuan Sun, and Li Sun. 2021. "A Crustin from Hydrothermal Vent Shrimp: Antimicrobial Activity and Mechanism" Marine Drugs 19, no. 3: 176. https://doi.org/10.3390/md19030176
APA StyleWang, Y., Zhang, J., Sun, Y., & Sun, L. (2021). A Crustin from Hydrothermal Vent Shrimp: Antimicrobial Activity and Mechanism. Marine Drugs, 19(3), 176. https://doi.org/10.3390/md19030176