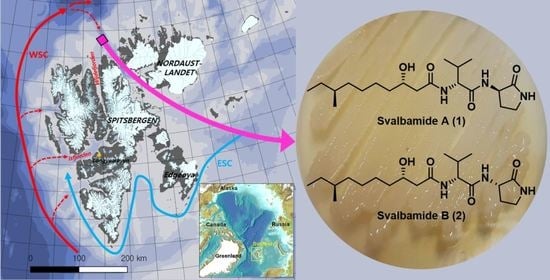
Svalbamides A and B, Pyrrolidinone-Bearing Lipodipeptides from Arctic Paenibacillus sp.
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phylogenetic Analysis
2.2. Structural Elucidation
2.3. Biological Evaluation
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Isolation, Cultivation, Phylogenetic Analysis, and Extraction of Bacteria
3.3. Isolation of Svalbamides A and B
3.4. Conformational Search and DP4 Analysis
3.5. Quinone Reductase Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.; An, J.S.; Bae, E.S.; Oh, J.; Park, S.H.; Lim, Y.; Ban, Y.H.; Kwon, Y.; Cho, J.-C.; Yoon, Y.J.; et al. Donghaesulfins A and B, dimeric benz[a]anthracene thioethers from volcanic island derived Streptomyces sp. Org. Lett. 2019, 21, 3635–3639. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.; An, J.S.; Hong, S.-H.; Bae, E.S.; Chung, B.; Kwon, Y.; Hong, S.; Oh, K.-B.; Shin, J.; Lee, S.K.; et al. Donghaecyclinones A–C: New cytotoxic rearranged angucyclinones from a volcanic island-derived marine Streptomyces sp. Mar. Drugs 2020, 18, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, K.; Ahn, C.-H.; Shin, Y.; Won, T.H.; Ko, K.; Lee, S.K.; Oh, K.-B.; Shin, J.; Nam, S.-I.; Oh, D.-C. New benzoxazine secondary metabolites from an arctic actinomycete. Mar. Drugs 2014, 12, 2526–2538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.K.; Bach, D.-H.; Ryu, H.W.; Oh, J.; Park, H.J.; Hong, J.-Y.; Song, H.-H.; Eum, S.; Bach, T.T.; Lee, S.K. Cytotoxic activities of Telectadium dongnaiense and its constituents by inhibition of the Wnt/β-catenin signaling pathway. Phytomedicine 2017, 34, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.S.; Lee, J.Y.; Kim, E.; Ahn, H.; Jang, Y.-J.; Shin, B.; Hwang, S.; Shin, J.; Yoon, Y.J.; Lee, S.K.; et al. Formicolides A and B, antioxidative and antiangiogenic 20-membered macrolides from a wood ant gut bacterium. J. Nat. Prod. 2020, 83, 2776–2784. [Google Scholar] [CrossRef] [PubMed]
- Cuendet, M.; Oteham, C.P.; Moon, R.C.; Pezzuto, J.M. Quinone reductase induction as a biomarker for cancer chemoprevention. J. Nat. Prod. 2006, 69, 460–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, K.C.-C.; Cao, S.; Raveh, A.; MacArthur, R.; Dranchak, P.; Chlipala, G.; Okoneski, M.T.; Guha, R.; Eastman, R.T.; Yuan, J.; et al. Actinoramide A identified as a potent antimalarial from titration-based screening of marine natural product extracts. J. Nat. Prod. 2015, 78, 2411–2422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoji, J.I.; Hinoo, H.; Sakazaki, R.; Kato, T.; Wakisaka, Y.; Mayama, M.; Matsuura, S.; Miwa, H. Isolation of tridecaptins A, B and C studies on antibiotics from the genus Bacillus. XXIII. J. Antibiot. 1978, 31, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, J.; Jukneviciute, G.; Čepaitė, R.; Vickackaite, V.; Pranckutė, R.; Kuisiene, N. Genome mining and characterization of biosynthetic gene clusters in two cave strains of Paenibacillus sp. Front. Microbiol. 2021, 11, 3433. [Google Scholar] [CrossRef] [PubMed]
- Bann, S.J.; Ballantine, R.D.; Cochrane, S.A. The tridecaptins: Non-ribosomal peptides that selectively target Gram-negative bacteria. RSC Med. Chem. 2021. [Google Scholar] [CrossRef]
- Cochrane, S.A.; Lohans, C.T.; van Belkum, M.J.; Bels, M.A.; Vederas, J.C. Studies on tridecaptin B1, a lipopeptide with activity against multidrug resistant gram-negative bacteria. Org. Biomol. Chem. 2015, 13, 6073–6081. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Shoji, J.I. The structure of octapeptin D studies on antibiotics from the genus Bacillus. XXVIII. J. Antibiot. 1980, 33, 186–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoji, J.I.; Kato, T.; Terabe, S.; Konaka, R. Resolution of peptide antibiotics, cerexins and tridecaptins, by high performance liquid chromatography studies on antibiotics from the genus Bacillus. XXVI. J. Antibiot. 1979, 32, 313–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Svalbamide A (1) a | Svalbamide B (2) a | |||||
|---|---|---|---|---|---|---|
| Position | δC, Type | δH, Mult (J in Hz) | δC, Type | δH, Mult (J in Hz) | ||
| 3-amino-2-pyrrolidinone | 1 | 174.2, C | 174.2, C | |||
| 2 | 49.4, CH | 4.30, m | 49.5, CH | 4.27, m | ||
| 3a | 28.0, CH2 | 1.82, m | 28.3, CH2 | 1.76, m | ||
| 3b | 2.26, m | 2.29, m | ||||
| 4 | 38.0, CH2 | 3.16, m | 38.0, CH2 | 3.16, m | ||
| 4-NH | 7.78, br s | 7.81, br s | ||||
| 2-NH | 8.10, d (8.5) | 8.21, d (8.5) | ||||
| d-Val | 5 | 171.0, C | 171.0, C | |||
| 6 | 57.2, CH | 4.18, dd (9.0, 6.5) | 57.2, CH | 4.20, dd (9.0, 6.5) | ||
| 7 | 30.7, CH | 1.96, m | 30.6, CH | 1.94, m | ||
| 8 | 18.0, CH3 | 0.84, d (7.0) | 18.0, CH3 | 0.83, d (7.0) | ||
| 9 | 19.3, CH3 | 0.88, d (7.0) | 19.1, CH3 | 0.84, d (7.0) | ||
| 6-NH | 7.83, d (9.0) | 7.85, d (9.0) | ||||
| 3-hydroxy-8-methyldecanoic acid | 10 | 170.8, C | 170.8, C | |||
| 11a | 43.4, CH2 | 2.23, dd (14.0, 7.0) | 43.4, CH2 | 2.25, dd (14.0, 7.0) | ||
| 11b | 2.29, dd (14.0, 5.0) | 2.28 dd (14.0, 5.0) | ||||
| 12 | 67.5, CH | 3.78, m | 67.6, CH | 3.78, m | ||
| 13 | 36.7, CH2 | 1.33, m b | 36.7, CH2 | 1.33, m b | ||
| 14a | 25.2, CH2 | 1.24, m b | 25.2, CH2 | 1.24, m b | ||
| 14b | 1.34, m b | 1.34, m b | ||||
| 15 | 26.5, CH2 | 1.23, m b | 26.5, CH2 | 1.23, m b | ||
| 16a | 36.0, CH2 | 1.05, m | 36.0, CH2 | 1.05, m | ||
| 16b | 1.25, m b | 1.25, m b | ||||
| 17 | 33.7, CH | 1.28, m b | 33.7, CH | 1.28, m b | ||
| 18a | 28.9, CH2 | 1.09, m | 28.9, CH2 | 1.09, m | ||
| 18b | 1.28, m b | 1.28, m b | ||||
| 19 | 11.2, CH3 | 0.82, t (7.0) | 11.2, CH3 | 0.83, t (7.0) | ||
| 20 | 19.1, CH3 | 0.81, d (6.5) | 19.1, CH3 | 0.81, d (6.5) | ||
| 12-OH | 4.65, d (5.0) | 4.67, d (5.0) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, Y.E.; Bae, E.S.; Lim, Y.; Cho, J.-C.; Nam, S.-J.; Shin, J.; Lee, S.K.; Nam, S.-I.; Oh, D.-C. Svalbamides A and B, Pyrrolidinone-Bearing Lipodipeptides from Arctic Paenibacillus sp. Mar. Drugs 2021, 19, 229. https://doi.org/10.3390/md19040229
Du YE, Bae ES, Lim Y, Cho J-C, Nam S-J, Shin J, Lee SK, Nam S-I, Oh D-C. Svalbamides A and B, Pyrrolidinone-Bearing Lipodipeptides from Arctic Paenibacillus sp. Marine Drugs. 2021; 19(4):229. https://doi.org/10.3390/md19040229
Chicago/Turabian StyleDu, Young Eun, Eun Seo Bae, Yeonjung Lim, Jang-Cheon Cho, Sang-Jip Nam, Jongheon Shin, Sang Kook Lee, Seung-Il Nam, and Dong-Chan Oh. 2021. "Svalbamides A and B, Pyrrolidinone-Bearing Lipodipeptides from Arctic Paenibacillus sp." Marine Drugs 19, no. 4: 229. https://doi.org/10.3390/md19040229
APA StyleDu, Y. E., Bae, E. S., Lim, Y., Cho, J. -C., Nam, S. -J., Shin, J., Lee, S. K., Nam, S. -I., & Oh, D. -C. (2021). Svalbamides A and B, Pyrrolidinone-Bearing Lipodipeptides from Arctic Paenibacillus sp. Marine Drugs, 19(4), 229. https://doi.org/10.3390/md19040229