Chemical Constituents of the Deep-Sea-Derived Penicillium citreonigrum MCCC 3A00169 and Their Antiproliferative Effects
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
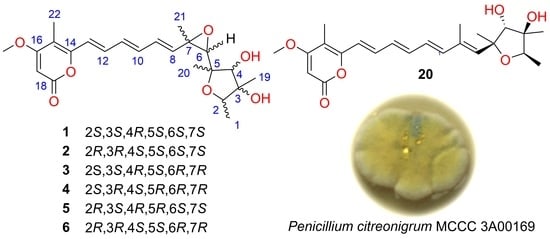
3.2. Biological Material
3.3. Fermentation and Extraction
3.4. Isolation and Purification
3.5. ECD Calculation
3.6. Measurement of ICD Spectra
3.7. The Antiproliferative Bioassay
3.8. Flow Cytometry
3.9. Western Blot Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Uchiyama, Y.; Takino, M.; Noguchi, M.; Shiratori, N.; Kobayashi, N.; Sugita-Konishi, Y. The In vivo and in vitro toxicokinetics of citreoviridin extracted from Penicillium citreonigrum. Toxins 2019, 11, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Sena Filho, J.G.; Hoover, A.R.; King, J.B.; Ellis, T.K.; Powell, D.R.; Cichewicz, R.H. Chemical epigenetics alters the secondary metabolite composition of guttate excreted by an atlantic-forest-soil-derived Penicillium citreonigrum. J. Nat. Prod. 2010, 73, 942–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, W.H.; Wei, Z.W.; Dai, P.; Wu, H.; Zhao, Y.X.; Zhang, M.M.; Jiang, N.; Zheng, W.F. Halogenated metabolites isolated from Penicillium citreonigrum. Chem. Biodivers. 2014, 11, 1078–1087. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.N.; Zou, Q.B.; Chen, J.; Xu, S.H.; Luo, D.; Zhang, F.G.; Lu, Y.Y. Phenols and diketopiperazines isolated from antarctic-derived fungi, Penicillium citreonigrum SP-6. Phytochem. Lett. 2018, 27, 114–118. [Google Scholar] [CrossRef]
- Tang, X.X.; Liu, S.Z.; Yan, X.; Tang, B.W.; Fang, M.J.; Wang, X.M.; Wu, Z.; Qiu, Y.K. Two new cytotoxic compounds from a deep-sea Penicillum citreonigrum XT20-134. Mar. Drugs 2019, 17, 509. [Google Scholar] [CrossRef] [Green Version]
- Yuan, W.H.; Goto, M.; Hsieh, K.Y.; Yuan, B.; Zhao, Y.; Morris-Natschke, S.L.; Lee, K.H. Selective cytotoxic eremophilane-type sesquiterpenes from Penicillium citreonigrum. J. Asian Nat. Prod. Res. 2015, 17, 1239–1244. [Google Scholar] [CrossRef] [Green Version]
- Yuan, W.H.; Zhang, Y.; Zhang, P.; Ding, R.R. Antioxidant sesquiterpenes from Penicillium citreonigrum. Nat. Prod. Commun. 2017, 12, 1827–1829. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.X.; Mudassir, S.; Zhang, Z.Z.; Feng, Y.Y.; Chang, Y.M.; Che, Q.; Gu, Q.Q.; Zhu, T.J.; Zhang, G.J.; Li, D.H. Secondary metabolites from deep-sea derived microorganisms. Curr. Med. Chem. 2020, 27, 6244–6273. [Google Scholar] [CrossRef]
- Xie, C.L.; Zhang, D.; Guo, K.Q.; Yan, Q.X.; Zou, Z.B.; He, Z.H.; Wu, Z.; Zhang, X.K.; Chen, H.F.; Yang, X.W. Meroterpenthiazole A, a unique meroterpenoid from the deep-sea-derived Penicillium allii-sativi, significantly inhibited retinoid X receptor (RXR)-α transcriptional effect. Chin. Chem. Lett. 2022, 33, 2057–2059. [Google Scholar] [CrossRef]
- Niu, S.; Xie, C.L.; Xia, J.M.; Liu, Q.M.; Peng, G.; Liu, G.M.; Yang, X.W. Botryotins A–H, tetracyclic diterpenoids representing three carbon skeletons from a deep-sea-derived Botryotinia fuckeliana. Org. Lett. 2020, 22, 580–583. [Google Scholar] [CrossRef]
- Niu, S.; Xia, J.M.; Li, Z.; Yang, L.H.; Yi, Z.W.; Xie, C.L.; Peng, G.; Luo, Z.H.; Shao, Z.; Yang, X.W. Aphidicolin chemistry of the deep-sea-derived fungus Botryotinia fuckeliana MCCC 3A00494. J. Nat. Prod. 2019, 82, 2307–2331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Mao, L.L.; Qian, P.T.; Shan, W.G.; Wang, J.D.; Bai, H. Two new metabolites from a soil fungus Curvularia affinis strain HS-FG-196. J. Asian Nat. Prod. Res. 2012, 14, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- Koga, J.; Yamauchi, T.; Shimura, M.; Ogawa, N.; Oshima, K.; Umemura, K.; Kikuchi, M.; Ogasawara, N. Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. J. Biol. Chem. 1998, 273, 31985–31991. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.H.; Zhang, Y.T.; Duan, D.; Fu, Q.; Zhang, X.Y.; Yu, X.W.; Wang, S.J.; Bao, B.; Wu, W.H. Fibrinolytic evaluation of compounds isolated from a marine fungus Stachybotrys longispora FG216. Chin. J. Chem. 2016, 34, 1194–1198. [Google Scholar] [CrossRef]
- Zhao, D.; Zheng, L.; Qi, L.; Wang, S.; Guan, L.; Xia, Y.; Cai, J. Structural features and potent antidepressant effects of total sterols and beta-sitosterol extracted from Sargassum horneri. Mar. Drugs 2016, 14, 123. [Google Scholar] [CrossRef] [PubMed]
- Uegaki, R.; Fujimori, T.; Kaneko, H.; Kato, K.; Noguchi, M. Isolation of dehydrololiolide and 3-oxo-actinidol from Nicotiana tabacum. Agric. Biol. Chem. 1979, 43, 1149–1150. [Google Scholar] [CrossRef] [Green Version]
- Tanimoto, S.; Tominaga, H.; Okada, Y.; Nomura, M. Synthesis and cosmetic whitening effect of glycosides derived from several phenylpropanoids. Yakugaku. Zasshi. 2006, 126, 173–177. [Google Scholar] [CrossRef] [Green Version]
- Arakawa, M.; Someno, T.; Kawada, M.; Ikeda, D. A new terrein glucoside, a novel inhibitor of angiogenin secretion in tumor angiogenesis. J. Antibiot. 2008, 61, 442–448. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.Y.; Wang, B.G.; Brauers, G.; Guan, H.S.; Proksch, P.; Ebel, R. Microsphaerones A and B, two novel gamma-pyrone derivatives from the sponge-derived fungus Microsphaeropsis sp. J. Nat. Prod. 2002, 65, 772–775. [Google Scholar] [CrossRef]
- You, J.; Yang, S.Z.; Mu, B.Z. Structural characterization of lipopeptides from Enterobacter sp strain N18 reveals production of surfactin homologues. Eur. J. Lipid Sci. Technol. 2015, 117, 890–898. [Google Scholar] [CrossRef]
- Nagia, M.M.; El-Metwally, M.M.; Shaaban, M.; El-Zalabani, S.M.; Hanna, A.G. Four butyrolactones and diverse bioactive secondary metabolites from terrestrial Aspergillus flavipes MM2: Isolation and structure determination. Org. Med. Chem. Lett. 2012, 2, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.Y.; Zhang, Y.; Yao, Y.B.; Lei, X.L.; Qian, Z.J. Butyrolactone-I from coral-derived fungus Aspergillus terreus attenuates neuro-inflammatory response via suppression of NF-kappaB Pathway in BV-2 Cells. Mar. Drugs 2018, 16, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Rocha, M.W.; Resck, I.S.; Caldas, E.D. Purification and full characterisation of citreoviridin produced by Penicillium citreonigrum in yeast extract sucrose (YES) medium. Food Addit. Contam. Part A 2015, 32, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Li, G.; Zhu, R.; Xie, F.; Li, Y.; Yang, W.; Xu, L.; Shen, T.; Zhao, Z.; Lou, H. Polyketides from the endolichenic fungus Eupenicillium javanicum and their anti-inflammatory activities. Phytochemistry 2020, 170, 112191. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.; Jiang, C.S.; Li, G.; Guo, Y.W. (+)-Cyclopenol, a new naturally occurring 7-membered 2,5-dioxopiperazine alkaloid from the fungus Penicillium sclerotiorum endogenous with the Chinese mangrove Bruguiera gymnorrhiza. J. Asian Nat. Prod. Res. 2014, 16, 542–548. [Google Scholar] [CrossRef]
- Campos, P.E.; Pichon, E.; Moriou, C.; Clerc, P.; Trepos, R.; Frederich, M.; De Voogd, N.; Hellio, C.; Gauvin-Bialecki, A.; Al-Mourabit, A. New antimalarial and antimicrobial tryptamine derivatives from the marine sponge Fascaplysinopsis reticulata. Mar. Drugs 2019, 17, 167. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.Y.; Liu, Y.Z.; Xu, J.; Jiang, F.W.; Kang, C.M. Synthesis of novel indole-benzimidazole derivatives. J. Chem. Res. 2016, 40, 588–590. [Google Scholar] [CrossRef]
- Hashida, J.; Niitsuma, M.; Iwatsuki, M.; Mori, M.; Ishiyama, A.; Namatame, M.; Nishihara-Tsukashima, A.; Nonaka, K.; Ui, H.; Masuma, R.; et al. Pyrenocine I, a new pyrenocine analog produced by Paecilomyces sp. FKI-3573. J. Antibiot. 2010, 63, 559–561. [Google Scholar] [CrossRef] [Green Version]
- Nakada, T.; Sudo, S.; Kosemura, S.; Yamamura, S. Two new metabolites of hybrid strains KO 0201 and 0211 derived from Penicillium citreoviride B. IFO 6200 and 4692. Tetrahedron Lett. 1999, 40, 6831–6834. [Google Scholar] [CrossRef]
- Hallock, Y.F.; Clardy, J.; Kenfield, D.S.; Strobel, G. De-O-methyldiaporthin, a phytotoxin from Drechslera siccans. Phytochemistry 1988, 27, 3123–3125. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Masuma, R.; Kim, Y.-P.; Uchida, R.; Tomoda, H.; Omura, S. Taxonomy and secondary metabolites of Pseudobotrytis sp. FKA-25. Mycoscience 2004, 45, 9–16. [Google Scholar] [CrossRef]
- Chen, C.; Ye, G.; Tang, J.; Li, J.; Liu, W.; Wu, L.; Long, Y. New polyketides from mangrove endophytic fungus Penicillium sp. BJR-P2 and their anti-inflammatory activity. Mar. Drugs 2022, 20, 583. [Google Scholar] [CrossRef] [PubMed]
- Frelek, J.; Ikekawa, N.; Takatsuto, S.; Snatzke, G. Application of [Mo2(OAc)4] for determination of absolute configuration of brassinosteroid vic-diols by circular dichroism. Chirality 1997, 9, 578–582. [Google Scholar] [CrossRef]
- Frelek, J.; Klimek, A.; Ruskowska, P. Dinuclear transition metal complexes as auxiliary chromophores in chiroptical studies on bioactive compounds. Curr. Org. Chem. 2003, 7, 1081–1104. [Google Scholar] [CrossRef]
- Frelek, J.; Pakulski, Z.; Zamojski, A. Application of [Mo2(OAc)4] for determination of absolute configuration of pyranoid and furanoid vic-diols by circular dichroism. Tetrahedron Asymmetry 1996, 7, 1363–1372. [Google Scholar] [CrossRef]
- Górecki, M.; Jabłońska, E.; Kruszewska, A.; Suszczyńska, A.; Urbańczyk-Lipkowska, Z.; Gerards, M.; Morzycki, J.W.; Szczepek, W.J.; Frelek, J. Practical method for the absolute configuration assignment of tert/tert 1,2-diols using their complexes with Mo2(OAc)4. J. Org. Chem. 2007, 72, 2906–2916. [Google Scholar] [CrossRef]
- Hou, H.; Zhou, R.; Li, A.; Li, C.; Li, Q.; Liu, J.; Jiang, B. Citreoviridin inhibits cell proliferation and enhances apoptosis of human umbilical vein endothelial cells. Environ. Toxicol. Pharmacol. 2014, 37, 828–836. [Google Scholar] [CrossRef]
- Myobatake, Y.; Kamisuki, S.; Tsukuda, S.; Higashi, T.; Chinen, T.; Takemoto, K.; Hachisuka, M.; Suzuki, Y.; Takei, M.; Tsurukawa, Y.; et al. Pyrenocine A induces monopolar spindle formation and suppresses proliferation of cancer cells. Bioorg. Med. Chem. 2019, 27, 115149. [Google Scholar] [CrossRef]
- Buachan, P.; Namsa-Aid, M.; Sung, H.K.; Peng, C.; Sweeney, G.; Tanechpongtamb, W. Inhibitory effects of terrein on lung cancer cell metastasis and angiogenesis. Oncol. Rep. 2021, 45, 94. [Google Scholar] [CrossRef]
- Feng, C.; Li, D.; Chen, M.; Jiang, L.; Liu, X.; Li, Q.; Geng, C.; Sun, X.; Yang, G.; Zhang, L.; et al. Citreoviridin induces myocardial apoptosis through PPAR-gamma-mTORC2-mediated autophagic pathway and the protective effect of thiamine and selenium. Chem. Biol. Interact. 2019, 311, 108795. [Google Scholar] [CrossRef]
- Zou, Z.B.; Zhang, G.; Li, S.M.; He, Z.H.; Yan, Q.X.; Lin, Y.K.; Xie, C.L.; Xia, J.M.; Luo, Z.H.; Luo, L.Z.; et al. Asperochratides A-J, ten new polyketides from the deep-sea-derived Aspergillus ochraceus. Bioorg. Chem. 2020, 105, 104349. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.X.; Yan, Q.X.; He, Z.H.; Zou, Z.B.; Le, Q.Q.; Chen, T.T.; Cai, B.; Yang, X.W.; Luo, S.L. Total synthesis and anti-inflammatory bioactivity of (−)-majusculoic acid and its derivatives. Mar. Drugs 2021, 19, 288. [Google Scholar] [CrossRef] [PubMed]









| No. | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 1.15 d (6.4) | 1.17 d (6.4) | 1.33 d (6.4) | 1.19 d (6.4) a | 1.20 d (6.4) | 1.19 d (6.4) |
| 2 | 3.92 q (6.4) a | 4.05 q (6.4) | 4.27 q (6.4) a | 4.10 q (6.4) | 4.09 q (6.4) a | 4.04 q (6.4) a |
| 4 | 4.06 s | 3.98 s | 3.76 s | 3.63 s | 4.02 s | 3.78 s |
| 6 | 3.60 s | 3.61 s | 3.81 s | 3.50 s | 3.63 s | 3.67 s |
| 8 | 6.41 d (15.2) | 6.06 d (15.2) | 5.96 d (15.2) | 6.56 d (15.2) | 6.01 d (15.2) | 6.02 d (15.2) |
| 9 | 6.51 dd (14.8, 10.8) | 6.42 dd (14.8, 10.8) a | 6.57 a | 6.38 dd (15.2, 10.8) | 6.42 dd (15.2, 10.8) a | 6.41 dd (15.2, 10.8) a |
| 10 | 6.54 dd (14.8, 10.8) | 6.51 dd (14.8, 10.8) a | 6.58 a | 6.61 dd (14.8, 10.8) a | 6.60 dd (14.8, 10.8) a | 6.64 dd (15.2, 10.8) a |
| 11 | 6.47 dd (14.8, 10.8) a | 6.48 dd (15.2, 10.8) | 6.48 a | 6.46 dd (14.8, 10.8) a | 6.48 dd (14.8, 10.8) a | 6.50 dd (14.8, 11.2) a |
| 12 | 7.14 dd (14.8, 10.8) | 7.13 dd (15.2, 10.8) | 7.12 dd (14.8, 10.8) | 7.14 dd (14.8, 10.8) | 7.14 dd (14.8, 10.8) | 7.16 dd (15.2, 10.8) |
| 13 | 6.49 d (15.2) | 6.55 d (15.2) | 6.60 d (14.8) a | 6.57 d (15.2) a | 6.59 d (15.2) a | 6.60 d (15.2) a |
| 17 | 5.62 s | 5.62 s | 5.62 s | 5.62 s | 5.63 s | 5.63 s |
| 19 | 1.29 s | 1.28 s | 1.21 s | 1.17 s | 1.18 s | 1.28 s |
| 20 | 1.23 s | 1.25 s | 1.21 s | 1.30 s | 1.27 s | 1.20 s |
| 21 | 1.35 s | 1.30 s | 1.30 s | 1.45 s | 1.29 s | 1.31 s |
| 22 | 2.00 s | 2.00 s | 2.00 s | 2.00 s | 2.01 s | 2.01 s |
| OMe | 3.90 s | 3.90 s | 3.91 s | 3.91 s | 3.90 s | 3.91 s |
| No. | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 13.6 q | 13.5 q | 12.1 q | 13.5 q | 13.8 q | 13.8 q |
| 2 | 79.2 d | 80.9 d | 81.9 d | 80.2 d | 80.0 d | 79.8 d |
| 3 | 84.1 s | 84.5 s | 85.2 s | 86.0 s | 84.9 s | 84.3 s |
| 4 | 74.8 d | 80.9 d | 81.7 d | 79.3 d | 79.4 d | 79.1 d |
| 5 | 84.3 s | 84.8 s | 87.4 s | 85.1 s | 87.5 s | 87.3 s |
| 6 | 84.1 d | 76.5 d | 85.5 d | 82.2 d | 91.0 d | 92.0 d |
| 7 | 77.2 s | 79.8 s | 76.9 s | 79.7 s | 74.3 s | 74.3 s |
| 8 | 144.7 d | 148.5 d | 144.0 d | 143.6 d | 143.5 d | 141.9 d |
| 9 | 129.0 d | 128.7 d | 129.4 d | 129.1 d | 129.5 d | 129.4 d |
| 10 | 139.7 d | 138.9 d | 138.8 d | 140.0 d | 139.1 d | 139.1 d |
| 11 | 131.7 d | 132.5 d | 132.6 d | 132.0 d | 132.5 d | 132.4 d |
| 12 | 137.3 d | 137.0 d | 137.1 d | 137.3 d | 137.1 d | 137.1 d |
| 13 | 120.0 d | 120.4 d | 120.4 d | 120.0 d | 120.4 d | 120.3 d |
| 14 | 156.0 s | 155.9 s | 155.9 s | 156.0 s | 155.9 s | 155.9 s |
| 15 | 109.6 s | 109.8 s | 109.8 s | 109.6 s | 109.8 s | 109.8 s |
| 16 | 173.2 s | 173.1 s | 173.1 s | 173.1 s | 173.1 s | 173.1 s |
| 17 | 89.0 d | 89.1 d | 89.1 d | 89.0 d | 89.1 d | 89.1 d |
| 18 | 166.4 s | 166.4 s | 166.4 s | 166.4 s | 166.4 s | 166.4 s |
| 19 | 17.5 q | 17.5 q | 13.1 q | 17.3 q | 13.2 q | 13.1 q |
| 20 | 16.6 q | 18.7 q | 14.4 q | 16.7 q | 14.6 q | 14.6 q |
| 21 | 32.2 q | 26.7 q | 26.6 q | 31.2 q | 26.2 q | 27.3 q |
| 22 | 8.9 q | 8.9 q | 8.9 q | 8.9 q | 8.9 q | 8.9 q |
| OMe | 57.3 q | 57.3 q | 57.3 q | 57.3 q | 57.3 q | 57.3 q |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, Z.-B.; Zhang, G.; Zhou, Y.-Q.; Xie, C.-L.; Xie, M.-M.; Xu, L.; Hao, Y.-J.; Luo, L.-Z.; Zhang, X.-K.; Yang, X.-W.; et al. Chemical Constituents of the Deep-Sea-Derived Penicillium citreonigrum MCCC 3A00169 and Their Antiproliferative Effects. Mar. Drugs 2022, 20, 736. https://doi.org/10.3390/md20120736
Zou Z-B, Zhang G, Zhou Y-Q, Xie C-L, Xie M-M, Xu L, Hao Y-J, Luo L-Z, Zhang X-K, Yang X-W, et al. Chemical Constituents of the Deep-Sea-Derived Penicillium citreonigrum MCCC 3A00169 and Their Antiproliferative Effects. Marine Drugs. 2022; 20(12):736. https://doi.org/10.3390/md20120736
Chicago/Turabian StyleZou, Zheng-Biao, Gang Zhang, Yu-Qi Zhou, Chun-Lan Xie, Ming-Min Xie, Lin Xu, You-Jia Hao, Lian-Zhong Luo, Xiao-Kun Zhang, Xian-Wen Yang, and et al. 2022. "Chemical Constituents of the Deep-Sea-Derived Penicillium citreonigrum MCCC 3A00169 and Their Antiproliferative Effects" Marine Drugs 20, no. 12: 736. https://doi.org/10.3390/md20120736
APA StyleZou, Z. -B., Zhang, G., Zhou, Y. -Q., Xie, C. -L., Xie, M. -M., Xu, L., Hao, Y. -J., Luo, L. -Z., Zhang, X. -K., Yang, X. -W., & Wang, J. -S. (2022). Chemical Constituents of the Deep-Sea-Derived Penicillium citreonigrum MCCC 3A00169 and Their Antiproliferative Effects. Marine Drugs, 20(12), 736. https://doi.org/10.3390/md20120736