Novel Prenylated Indole Alkaloids with Neuroprotection on SH-SY5Y Cells against Oxidative Stress Targeting Keap1–Nrf2
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemical Structure Elucidation
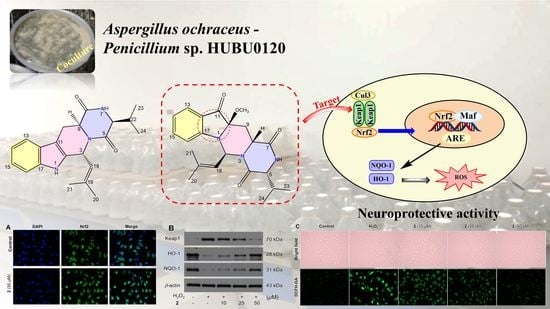
2.2. Neuroprotection on SH-SY5Y Cells against Oxidative Stress
2.3. Molecular Docking and Dynamics Simulation of 2–1X2R
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Strain Material
3.3. Fermentation, Extraction and Isolation
3.4. Single-Crystal X-ray Data for Asperpenazine (1)
3.5. Cytotoxicity and Cytoprotection Evaluation
3.6. ROS Level Evaluation
3.7. GSH Level Evaluation
3.8. Nuclear Translocation of Nrf2
3.9. Western Blotting
3.10. Quantitative Real-Time Reverse Transcriptase—Polymerase Chain Reaction (qRT−PCR)
3.11. Molecular Docking
3.12. Molecular Dynamics Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DEPT | Distortionless enhancement by polarization transfer |
| HSQC | 1H detected heteronuclear single quantum coherence spectroscopy |
| HMBC | 1H detected heteronuclear multiple bond connectivity spectroscopy |
| 1H–1H COSY | 1H–1H chemical shift correlated spectroscopy |
| NOESY | Nuclear overhauser effect spectroscopy |
| ORTEP | Oak ridge thermal ellipsoid plot |
| CCK-8 | Cell counting kit-8 |
| DCFH–DA | 2,7-Dichlorodihydrofluorescein diacetate |
| ELISA | Enzyme-linked immunosorbent assay |
| DAPI | 4′,6-Diamidino-2-phenylindole |
References
- Gao, C.; Sun, H.; Wang, T.; Tang, M.; Bohnen, N.I.; Müller, M.L.; Herman, T.; Giladi, N.; Kalinin, A.; Spino, C.; et al. Model-based and model-free machine learning techniques for diagnostic prediction and classification of clinical outcomes in Parkinson’s disease. Sci. Rep. 2018, 8, 7129. [Google Scholar] [CrossRef]
- Nikam, S.; Nikam, P.; Ahaley, S.K.; Sontakke, A.V. Oxidative stress in Parkinson’s disease. Indian J. Clin. Biochem. 2009, 24, 98–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thanan, R.; Oikawa, S.; Hiraku, Y.; Ohnishi, S.; Ma, N.; Pinlaor, S.; Yongvanit, P.; Kawanishi, S.; Murata, M. Oxidative stress and its significant roles in neurodegenerative diseases and cancer. Int. J. Mol. Sci. 2014, 16, 193–217. [Google Scholar] [CrossRef] [Green Version]
- Gan, L.; Johnson, J.A. Oxidative damage and the Nrf2-ARE pathway in neurodegenerative diseases. Biochim. Biophys. Acta 2014, 1842, 1208–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, M.M.; Rocha, C.R.R.; Kinker, G.S.; Pelegrini, A.L.; Menck, C.F.M. The balance between NRF2/GSH antioxidant mediated pathway and DNA repair modulates cisplatin resistance in lung cancer cells. Sci. Rep. 2019, 9, 17639. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.H.; Zhang, J.T.; Yang, G.J.; Liu, H.; Han, Q.B.; Ma, D.L. Emerging screening approaches in the development of Nrf2-Keap1 protein-protein interaction inhibitors. Int. J. Mol. Sci. 2019, 20, 4445. [Google Scholar] [CrossRef] [Green Version]
- Pallesen, J.S.; Tran, K.T.; Bach, A. Non-covalent small-molecule Kelch-like ECH-associated protein 1-nuclear factor erythroid 2-related factor 2 (Keap1-Nrf2) inhibitors and their potential for targeting central nervous system diseases. J. Med. Chem. 2018, 61, 8088–8103. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef]
- Schueffler, A.; Anke, T. Fungal natural products in research and development. Nat. Prod. Rep. 2014, 31, 1425–1448. [Google Scholar] [CrossRef]
- Finefield, J.M.; Frisvad, J.C.; Sherman, D.H.; Williams, R.M. Fungal origins of the bicyclo[2.2.2]diazaoctane ring system of prenylated indole alkaloids. J. Nat. Prod. 2012, 75, 812–833. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.M.; Stocking, E.M.; Sanz Cervera, J.F. Biosynthesis of prenylated alkaloids derived from Tryptophan. Topics Curr. Chem. 2000, 209, 97–173. [Google Scholar]
- Li, S.M. Genome mining and biosynthesis of fumitremorgin-type alkaloids in ascomycetes. J. Antibiot. 2011, 64, 45–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Wen, J.; Zhu, T.; Fang, Y.; Gu, Q.; Zhu, W. Chrysogenamide A from an endophytic fungus associated with Cistanche deserticola and its neuroprotective effect on SH-SY5Y cells. J. Antibiot. 2008, 61, 81–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Tian, S.; Wu, R.; Tong, Z.; Jiang, W.; Hu, P.; Xiao, X.; Zhang, X.; Zhou, H.; Tong, Q.; et al. Identification of anti-Parkinson’s disease lead compounds from Aspergillus ochraceus targeting adenosin receptors A2A. ChemistryOpen 2021, 10, 630–638. [Google Scholar] [CrossRef]
- Zhang, C.; Hu, L.; Liu, D.; Huang, J.; Lin, W. Circumdatin D exerts neuroprotective effects by attenuating LPS-induced pro-inflammatory responses and downregulating acetylcholinesterase activity in vitro and in vivo. Front. Pharmacol. 2020, 11, 760. [Google Scholar] [CrossRef]
- Tong, Z.; Xiao, X.; Lu, Y.; Zhang, Y.; Hu, P.; Jiang, W.; Zhou, H.; Pan, S.; Huang, Z.; Hu, L. New metabolites from Aspergillus ochraceus with antioxidative activity and neuroprotective potential on H2O2 insult SH-SY5Y cells. Molecules 2022, 27, 52. [Google Scholar] [CrossRef]
- Grundmann, A.; Kuznetsova, T.; Afiyatullov, S.; Li, S.M. FtmPT2, an N-prenyltransferase from Aspergillus fumigatus, catalyses the last step in the biosynthesis of fumitremorgin B. ChemBioChem 2008, 9, 2059–2063. [Google Scholar] [CrossRef]
- Xie, F.; Li, X.B.; Zhou, J.C.; Xu, Q.Q.; Wang, X.N.; Yuan, H.Q.; Lou, H.X. Secondary metabolites from Aspergillus fumigatus, an endophytic fungus from the Liverwort Heteroscyphus tener (Steph.) Schiffn. Chem. Biodivers. 2015, 12, 1313–1321. [Google Scholar] [CrossRef]
- He, X.; Ding, L.; Yi, M.; Xu, J.; Zhou, X.; Zhang, W.; He, S. Separation of five diketopiperazines from the marine fungus Alternaria alternate HK-25 by high-speed counter-current chromatography. J. Sep. Sci. 2019, 42, 2510–2516. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.K.; Wang, W.X. GIAO 13C NMR Calculation with sorted training sets improves accuracy and reliability for structural assignation. J. Org. Chem. 2020, 85, 11350–11358. [Google Scholar] [CrossRef]
- Lodewyk, M.W.; Siebert, M.R.; Tantillo, D.J. Computational prediction of 1H and 13C chemical shifts: A useful tool for natural product, mechanistic, and synthetic organic chemistry. Chem. Rev. 2012, 112, 1839–1862. [Google Scholar] [CrossRef] [PubMed]
- Maiya, S.; Grundmann, A.; Li, S.M.; Turner, G. The fumitremorgin gene cluster of Aspergillus fumigatus: Identification of a gene encoding brevianamide F synthetase. ChemBioChem 2006, 7, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Suzuki, H.; Takagi, H.; Asami, Y.; Kakeya, H.; Uramoto, M.; Usui, T.; Takahashi, S.; Sugimoto, Y.; Osada, H. Identification of cytochrome P450s required for fumitremorgin biosynthesis in Aspergillus fumigatus. ChemBioChem 2009, 10, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Liu, M.; Jenkins, I.D.; Liu, X.; Zhang, L.; Quinn, R.J.; Feng, Y. Genome-inspired chemical exploration of marine fungus Aspergillus fumigatus MF071. Mar. Drugs 2020, 18, 352. [Google Scholar] [CrossRef]
- Dolle, R.E. Comprehensive survey of chemical libraries yielding enzyme inhibitors, receptor agonists and antagonists, and other biologically active agents: 1992 through 1997. Mol. Divers. 1998, 3, 199–233. [Google Scholar] [CrossRef]
- Narasimhan, M.; Patel, D.; Vedpathak, D.; Rathinam, M.; Henderson, G.; Mahimainathan, L. Identification of novel microRNAs in post-transcriptional control of Nrf2 expression and redox homeostasis in neuronal, SH-SY5Y cells. PLoS ONE 2012, 7, e51111. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.A.; Gao, M.; Zhang, L.; Zhao, Y.; Shi, L.L.; Chen, B.N.; Wang, Y.H.; Wang, S.B.; Du, G.H. Salvianolic acid A protects human SH-SY5Y neuroblastoma cells against H2O2-induced injury by increasing stress tolerance ability. Biochem. Biophys. Res. Commun. 2012, 421, 479–483. [Google Scholar] [CrossRef]
- Heo, S.R.; Han, A.M.; Kwon, Y.K.; Joung, I. P62 protects SH-SY5Y neuroblastoma cells against H2O2-induced injury through the PDK1/Akt pathway. Neurosci. Lett. 2009, 450, 45–50. [Google Scholar] [CrossRef]
- González Reyes, S.; Guzmán Beltrán, S.; Medina Campos, O.N.; Pedraza Chaverri, J. Curcumin pretreatment induces Nrf2 and an antioxidant response and prevents hemin-induced toxicity in primary cultures of cerebellar granule neurons of rats. Oxid. Med. Cell. Longev. 2013, 2013, 801418. [Google Scholar] [CrossRef]
- Morroni, F.; Sita, G.; Graziosi, A.; Turrini, E.; Fimognari, C.; Tarozzi, A.; Hrelia, P. Protective effects of 6-(Methylsulfinyl)hexyl isothiocyanate on Aβ1-42-induced cognitive deficit, oxidative stress, inflammation, and apoptosis in mice. Int. J. Mol. Sci. 2018, 19, 2083. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Meng, N.; Liu, Z.; Guo, L.; Dong, L.; Li, B.; Ye, Q. Neuroprotective effects of Taraxacum officinale Wigg. extract on glutamate-induced oxidative stress in HT22 cells via HO-1/Nrf2 pathways. Nutrients 2018, 10, 926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.J.; Guo, C.L.; Li, X.Q.; Wang, S.Y.; Jiang, B.; Zhao, Y.; Luo, J.; Xu, K.; Liu, H.; Guo, S.J.; et al. Discovery of novel bromophenol hybrids as potential anticancer agents through the Ros-mediated apoptotic pathway: Design, synthesis and biological evaluation. Mar. Drugs 2017, 15, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, C.S.; Kim, D.C.; Lee, D.S.; Kim, K.S.; Ko, W.; Sohn, J.H.; Yim, J.H.; Kim, Y.C.; Oh, H. Anti-neuroinflammatory effect of aurantiamide acetate from the marine fungus Aspergillus sp. SF-5921: Inhibition of NF-κB and MAPK pathways in lipopolysaccharide-induced mouse BV2 microglial cells. Int. Immunopharmacol. 2014, 23, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Vainio, M.J.; Johnson, M.S. Generating conformer ensembles using a multiobjective genetic algorithm. J. Chem. Inf. Model 2007, 47, 2462–2474. [Google Scholar]
- O’Boyle, N.M.; Vandermeersch, T.; Flynn, C.J.; Maguire, A.R.; Hutchison, G.R. Confab-systematic generation of diverse low-energy conformers. J. Cheminform. 2011, 3, 1–9. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. (Eds.) Gaussian 16; Revision A.03; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]









| No. | 1 | 2 | ||
|---|---|---|---|---|
| δH | δC | δH | δC | |
| 2 | 132.6 | 7.23 d (9.8) | 58.5 | |
| 3 | 6.45 d (9.5) | 48.3 | ||
| 4 | 163.0 | |||
| 5 | 163.8 | 3.91 brt (2.4) | 60.1 | |
| 6 | 3.98 brt (2.4) | 60.9 | ||
| 7 | 166.6 | |||
| 8 | 168.4 | 4.66 dd (13.0, 2.3) | 48.9 | |
| 9 | 4.38 dd (11.7, 4.0) | 53.6 | Ha 1.53 t (13.2) Hb 2.64 dd (13.8, 2.6) | 36.4 |
| 10 | Ha 3.46 dd (15.4, 4.2) Hb 2.87 ddd (15.3, 11.8, 11.3) | 28.2 | 91.1 | |
| 11 | 106.6 | 198.4 | ||
| 12 | 126.7 | 119.6 | ||
| 13 | 7.42 d (7.7) | 118.4 | 7.59 d (8.0) | 125.5 |
| 14 | 7.06 td (7.6, 1.1) | 120.1 | 6.87 t (7.5) | 120.3 |
| 15 | 7.12 td (7.6, 1.1) | 122.5 | 7.55 d (7.7) | 138.7 |
| 16 | 7.25 d (7.9) | 111.2 | 7.17 d (8.4) | 110.7 |
| 17 | 136.3 | 157.7 | ||
| 18 | 5.27 dt (9.6, 1.3) | 120.7 | 5.77 d (8.6) | 117.9 |
| 19 | 139.0 | 139.4 | ||
| 20 | 1.71 d (1.1) | 26.2 | 1.79 s | 26.1 |
| 21 | 1.98 d (1.1) | 19.1 | 1.91 s | 18.8 |
| 22 | 2.38 ddt (10.5, 6.8, 3.7) | 32.7 | 2.29 ddt (13.7, 7.0, 3.2) | 32.2 |
| 23 | 0.86 d (6.8) | 19.0 | 0.88 d (7.1) | 18.7 |
| 24 | 1.02 d (7.0) | 16.6 | 0.52 d (6.8) | 16.0 |
| 25-OCH3 | 3.14 s | 52.0 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, X.; Tong, Z.; Zhang, Y.; Zhou, H.; Luo, M.; Hu, T.; Hu, P.; Kong, L.; Liu, Z.; Yu, C.; et al. Novel Prenylated Indole Alkaloids with Neuroprotection on SH-SY5Y Cells against Oxidative Stress Targeting Keap1–Nrf2. Mar. Drugs 2022, 20, 191. https://doi.org/10.3390/md20030191
Xiao X, Tong Z, Zhang Y, Zhou H, Luo M, Hu T, Hu P, Kong L, Liu Z, Yu C, et al. Novel Prenylated Indole Alkaloids with Neuroprotection on SH-SY5Y Cells against Oxidative Stress Targeting Keap1–Nrf2. Marine Drugs. 2022; 20(3):191. https://doi.org/10.3390/md20030191
Chicago/Turabian StyleXiao, Xueyang, Zhou Tong, Yuexing Zhang, Hui Zhou, Mengying Luo, Tianhui Hu, Ping Hu, Luqi Kong, Zeqin Liu, Chan Yu, and et al. 2022. "Novel Prenylated Indole Alkaloids with Neuroprotection on SH-SY5Y Cells against Oxidative Stress Targeting Keap1–Nrf2" Marine Drugs 20, no. 3: 191. https://doi.org/10.3390/md20030191
APA StyleXiao, X., Tong, Z., Zhang, Y., Zhou, H., Luo, M., Hu, T., Hu, P., Kong, L., Liu, Z., Yu, C., Huang, Z., & Hu, L. (2022). Novel Prenylated Indole Alkaloids with Neuroprotection on SH-SY5Y Cells against Oxidative Stress Targeting Keap1–Nrf2. Marine Drugs, 20(3), 191. https://doi.org/10.3390/md20030191